Electrochemical,spectroscopic,and molecular docking studies of the interaction between the anti-retroviral drug indinavir and dsDNA
Frib Mollrsouli,Burcu Dogn-Topl,Mehmet Gokhn Cglyn,Tugb Tskin-Tok,Sibel A.Ozkn,*
aAnkara University,Department of Analytical Chemistry,06560,Ankara,Turkey
bDepartment of Chemistry,Yasouj University,Yasouj,75918-74831,Iran
cDepartment of Chemistry,Gaziantep University,27310,Gaziantep,Turkey
dInstitute of Health Sciences,Department of Bioinformatics and Computational Biology,Gaziantep University,27310,Gaziantep,Turkey
Keywords:
Glassy carbon electrode
Calf thymus double-stranded
deoxyribonucleic acid(ct-dsDNA)
Biosensor
Fluorescence
Molecular docking
ABSTRACT
In this study,an electrochemical DNA biosensor was developed using a straightforward methodology to investigate the interaction of indinavir with calf thymus double-stranded deoxyribonucleic acid(ctdsDNA)for the first time.The decrease in the oxidation signals of deoxyguanosine(dGuo)and deoxyadenosine(dAdo),measured by differential pulse voltammetry,upon incubation with different concentrations of indinavir can be attributed to the binding mode of indinavir to ct-dsDNA.The currents of the dGuo and dAdo peaks decreased linearly with the concentration of indinavir in the range of 1.0-10.0 μg/mL.The limit of detection and limit of quantification for indinavir were 0.29 and 0.98 μg/mL,respectively,based on the dGuo signal,and 0.23 and 0.78μg/mL,respectively,based on the dAdo signal.To gain further insights into the interaction mechanism between indinavir and ct-dsDNA,spectroscopic measurements and molecular docking simulations were performed.The binding constant(Kb)between indinavir and ct-dsDNA was calculated to be 1.64× 108M-1,based on spectrofluorometric measurements.The obtained results can offer insights into the inhibitory activity of indinavir,which could help to broaden its applications.That is,indinavir can be used to inhibit other mechanisms and/or hallmarks of viral diseases.
1.Introduction
Indinavir(IND),made by Merck under the trading name of Crixivan?(Fig.1),is an anti-retroviral drug acting as a competitive and reversible protease inhibitor,and has an essential role against human immunodeficiency virus infection and acquired immune deficiency syndrome(HIV/AIDS)replication[1].It is a white powder soluble in water that can be administered orally.It may be used in combination with other medications,such as ritonavir,to boost IND activity during highly active anti-retroviral therapy(HAART)[2].IND does not affect the early stages of HIV replication;instead,it disrupts the production of infectious HIV,decreasing the contagiousness of the virus[3].Therefore,IND works better for controlling HIV infection.The mechanism of action of IND depends on the inhibition of the HIV protease,blocking virus maturation and decreasing the number of reproducing HIV viruses,thus causing a decrease in the viral load and the formation of immature and noninfectious virions.Hence,the immune system can work better.IND can act in both chronically and acutely infected cells,whereas nucleoside reverse transcriptase inhibitors(NRTIs)do not affect chronically infected cells.In commercially available IND,an additional amine in the hydroxy ethylene backbone of the anhydrous form of the drug is added to enhance solubility and oral bioavailability,facilitating drug intake.In vitro studies have shown that the antiviral effects of HIV protease inhibitors and those of non-nucleoside reverse transcriptase inhibitors(NNRTIs)or some NRTIs might be additive or synergistic[3].IND can be detected in the cerebrospinal fluid of a person who is administered the drug.Over a concentration range of 0.05-10 mg/mL,approximately 60% of IND can bind to proteins.IND is mainly metabolized by the liver and is rapidly absorbed,reaching its highest plasma concentration within0.8±0.3h.The average plasma half-life of IND is 1.8 h for adults and 1.1 h for children[3].
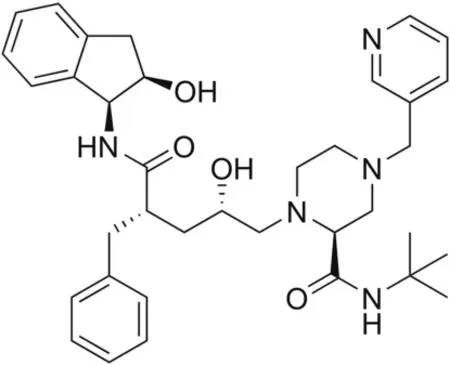
Fig.1.The structure of indinavir.
Several techniques,including UV absorbance measurements[4],high-performance liquid-chromatography(HPLC),electrochemistry[5-7],capillary electrophoresis[8],and liquid-chromatographytandem mass spectroscopy(LC-MS/MS)[9],have been used for IND detection.However,the availability of time-and cost-saving methods that can detect IND with high sensitivity opens new possibilities for the future of pharmaceutical analysis.Electrochemistry offers such advantages.In a study conducted by Dogan et al.[6]in 2006,the behavior of IND subjected to electrochemical reduction was investigated.The results indicated a diffusion controlled process,based on the IND peak.The limit of detection(LOD)was determined to be as low as 1.26 × 10-7M[6].To the best of our knowledge,the interaction between IND and dsDNA has not been studied by any method.The study of the drug-DNA interaction using electrochemistry can be used not only to understand the mechanism of drug action and to design new and more efficient drugs,but also to determine the specific drug compounds.
DNA exists in the form of a double helix,consisting of two strands composed of four types of nucleobase subunits:guanine(G),cytosine(C),adenine(A),and thymine(T)[10].The two strands are held together mainly through Watson Crick hydrogen bonds,with C-G pairs linked with three hydrogen bonds,and A-T pairs with two hydrogen bonds[11].
DNA replication and transcription are two major cell functions.DNA transcription is required for gene expression and protein synthesis,and therefore,plays key roles in health and disease.Some small organic molecules can mimic signaling regulatory proteins in terms of binding strength and specificity.Consequently,these molecules can activate DNA replication and/or transcription,and increase protein production.On the contrary,other small organic molecules can inhibit DNA-related processes,restricting DNA replication and protein synthesis,leading to cell apoptosis.In most cases,DNA-targeted drugs for antibiotic and antitumor applications are designed to inhibit DNA-related processes,leading to cell apoptosis.Small organic molecules can interact with DNA through groove binding,intercalation,and/or cross-linking(covalent bonding).Complex formation between small organic molecules and DNA through covalent binding or non-covalent interactions results in changes in the functional properties and thermal stability of DNA[12].Often,such changes have a great impact on physiological functions.Therefore,it is critical to understand the mechanisms underlying small molecule binding to DNA and to elucidate the functional properties of the resulting complexes,which may have applications for drug research and development[13].
Small organic molecules can bind at either the minor or major groove of DNA.The minor and major grooves are a result of the helical structure of the DNA.The glycosidic bonds in the two DNA strands are always in the same location within a base pair,and they are neither parallel nor perpendicular to one another.This asymmetric positioning of the glycosidic bonds creates the major groove at the side where the backbones of the two DNA strands are far apart,and the minor groove at the opposite site,where the backbones of the two strands come close together.In general,groove binding,contrary to intercalation,occurs in two steps.During the first step,the binding molecule undergoes a hydrophobic transfer from the solution to the groove of DNA.During the second step,the binding molecule forms non-covalent interactions with the DNA nucleobases.Groov ebinding does not require unwinding of the DNA helical axis.Typically,groove binders prefer to bind at AT-rich sequences because GC-rich sequences contain a-NH2group that sterically inhibits binding[12,13].
In this study,the interaction of IND with calf thymus doublestranded DNA(ct-dsDNA)was investigated with a glassy carbon electrode(GCE)modified with a multilayer of ct-dsDNA.The results were confirmed by three independent techniques,namely,electrochemical and fluorescence measurements,and molecular modeling.
2.Experimental
2.1.Apparatus and software
Electrochemical measurements were performed with an AUTOLAB PGSTAT 30 potentiostat/galvanostat(Eco Chemie,the Netherlands),equipped with a three-electrode cell system and running the GPES 4.9 software.The three-electrode system consisted of an Ag/AgCl electrode(BASi;3 M NaCl)as the reference electrode,a bare and modified GCE as the working electrode(φ=3.0 mm for the unmodified electrode shipped from BASi),and a platinum counter electrode.The pH was adjusted using a digital pH meter Model 538(WTW,Austria),equipped with a combined glass electrode.UV-vis absorption spectra were recorded on the Agilent Cary 60 UV-Vis Spectrophotometer.Spectrofluorometric measurements were conducted on the Agilent Cary Eclipse Fluorescence Spectrophotometer,using a quartz fluorescence cuvette(10 mm×10 mm).
2.2.Reagents and solutions
Indinavir sulfate(IND)was kindly provided by Merck Sharp&Dohme Pharm.Ind.(Istanbul,Turkey).An IND stock solution(1000μg/mL)was prepared by dissolving the powder in ultrapure water and was stored in the refrigerator.Acetic acid,sodium acetate,and methylene blue were purchased from Merck.Calf thymus double-stranded DNA(ct-dsDNA)was purchased from Sigma(D1501),and was used for studying its interaction with IND.The stock solution of ct-dsDNA(500μg/mL)was prepared in ultrapure water from a Millipore Milli-Q system and stored at 4°C overnight.Further required dilutions of the ct-dsDNA stock solution were prepared in ultrapurewater.Acetate buffer was prepared by mixing 0.1 M sodium acetate and 0.1 M acetic acid solutions.The concentration of the ct-dsDNA stock solution was determined by UV absorption measurements at 260 nm according to Lambert-Beer’s Law equation,A = εbc,where A represents the absorption value,ε is the molar absorption coefficient(ε260= 6600 L/mol/cm),b is the cuvette path length(1 cm),and c represents the drug concentration.To evaluate the purity of ct-dsDNA,the ratio of the absorbance at 260 nm to the absorbance at 280 nm(A260/A280)was calculated,and if it was higher than 1.8,it could be concluded that the ctdsDNA preparation was sufficiently protein-free[14].All chemicals used were of analytical reagent grade,and their solutions were prepared by using double distilled water.
2.3.Electrochemical measurements
Differential pulse(DP)voltammograms were recorded from 0.2 to 1.5 V vs.Ag/AgCl(3 M NaCl),using the following parameters:pulse amplitude=50 mV,pulse time=70 m s,and scan rate=5 mV/s.For the incubation in the solution phase,a mixture solution was prepared by mixing 100 μg/mL ct-dsDNA with 10 μg/mL IND in 0.1 M acetate buffer(pH=4.70),and then,incubated for different periods of time at room temperature.To calculate the binding constant,increasing concentrations of ct-dsDNA(4.83 × 10-6-1.60× 10-5M)were added to a fixed concentration of IND in solution.
To fabricate the DNA biosensor,the surface of the cleaned GCE was coated by drop-casting with 5 μL of 50 μg/mL ct-dsDNA(prepared freshly),followed by drying in an oven for 10 min at 45°C.This step was repeated three times for each biosensor.Finally,the prepared DNA biosensor was cleaned by immersion into a blank acetate buffer solution for 1 s to remove the unbound ct-dsDNA from the electrode surface.The ct-dsDNA-modified biosensor was immersed in an IND solution,and was incubated for different time periods.Then,the biosensor was cleaned again with blank acetate buffer solution for 5 s to remove any unbound IND[15].DP voltammograms were recorded in a cleaned blank acetate buffer solution(0.1 M,pH=4.70)in the range of 0.2 and 1.5 V vs.Ag/AgCl(3 M NaCl).Each experiment was performed with a new biosensor.
2.4.Fluorescence measurements
Fluorescence spectra of methylene blue(MB)at a concentration of 5.0μM were recorded in the absence or presence of different ctdsDNA concentrations with a spectrofluorometer using the following parameters:excitation wavelength=290 nm,excitation and emission slits=10 nm,photomultiplier tube voltage=675 V.The fluorescence spectra were recorded continually while adding dropwise small volumes(1μL)of a stock solution of 1.92 mM ctdsDNA to a 50μM aqueous solution of MB under stirring.Fluorescence measurements at different temperatures were recorded after waiting 3 min at each temperature to reach equilibrium.Millipore Milli-Q water was used as a blank solution.Progressive quenching of the MB fluorescence signal was observed at increasing DNA concentrations,indicating an interaction between MB and ct-dsDNA.It was assumed that the observed fluorescence quenching could be attributed to the formation of a fluorophorequencher complex.When the MB fluorescence signal did not change with the further addition of ct-dsDNA,we started to add small volumes(1μL)of a stock solution of 5 mM IND to the fluorophore-quencher complex solution.In this case,the MB fluorescence signal was recovered because of the stronger DNA binding properties of IND compared to MB,which resulted in the displacement of MB from the MB-DNA complex.This process was continued until a constant fluorescence signal for MB was reached.All fluorescence experiments were repeated at different temperatures(297 K,301 K,305 K,and 311 K)adjusted by a Peltier temperature controller.
2.5.Molecular modeling study
Molecular docking with IND as the ligand and DNA as the target was performed using the Discovery Studio(DS)2019 software suite[16]to investigate and visualize the interaction mechanisms and the possible orientations of the complex.The crystal structure of DNA(ID:1BNA)[17]was uploaded from the protein data bank,PDB(http://www.rcsb.org/pdb).Additionally,the three dimensional(3D)coordinates of IND were optimized at DFT/B3LYP/6-31G×level by using the Gaussian 09(G09)software[18].The optimized IND structure and all related information are given in Fig.S1 and Table S1.The target(DNA)was also minimized with the‘prepare protein’protocol of DS 2019[16].This protocol uses the CHARMM force field and the adopted-basis Newton-Raphson(ABNR)method[19]to prepare the target for molecular docking.At the end of the docking simulations,the best IND conformer was selected from 50 diversified conformers,based on high negative values of the calculated binding energy,three-dimensional interaction types,and the lowest values of root mean square deviation(RMSD),using DS 2019[16].
3.Results and discussion
3.1.Electrochemical studies of the IND-ct-dsDNA interaction
As seen in Fig.2,the DP voltammograms of 10μg/mL IND in 0.1 M acetate buffer solution(pH=4.70)in the potential range of 0.2-1.5 V vs.Ag/AgCl(3 M NaCl)show one oxidation peak at a potential of 0.80 V.
The molecular structure of IND reveals the presence of a piperazine ring in Ref.[24],which is a basic center that,owing to its available non-bonding electrons,can act as a donor.Therefore,it can be assumed that the main oxidation step of IND should occur on the piperazine ring.According to the literature,a nitrogen on the piperazine ring loses an electron to form a cation radical,which,in subsequent steps,loses a proton and an electron to form a quaternary Schiff base[25-27].
The DP voltammograms of the biosensor-attached DNA at the same electrolyte displayed two oxidation peaks,attributed to the oxidation of dGuo at 0.96 V and that of dAdo at 1.23 V vs.Ag/AgCl(3 M NaCl).It is clear from Fig.2 that no changes were observed when the DNA biosensor was incubated in the 0.1 M acetate buffer solution(pH=4.70)without the addition of the IND drug.On the contrary,a significant decrease in the current of the dGuo and dAdo peaks,along with a positive shift in the dGuo oxidation peak potential,was observed after incubating the DNA biosensor in a 0.1 M acetate buffer solution(pH=4.70)containing the IND drug,indicating possible interactions between IND and ct-dsDNA.
3.1.1.In-solution phase
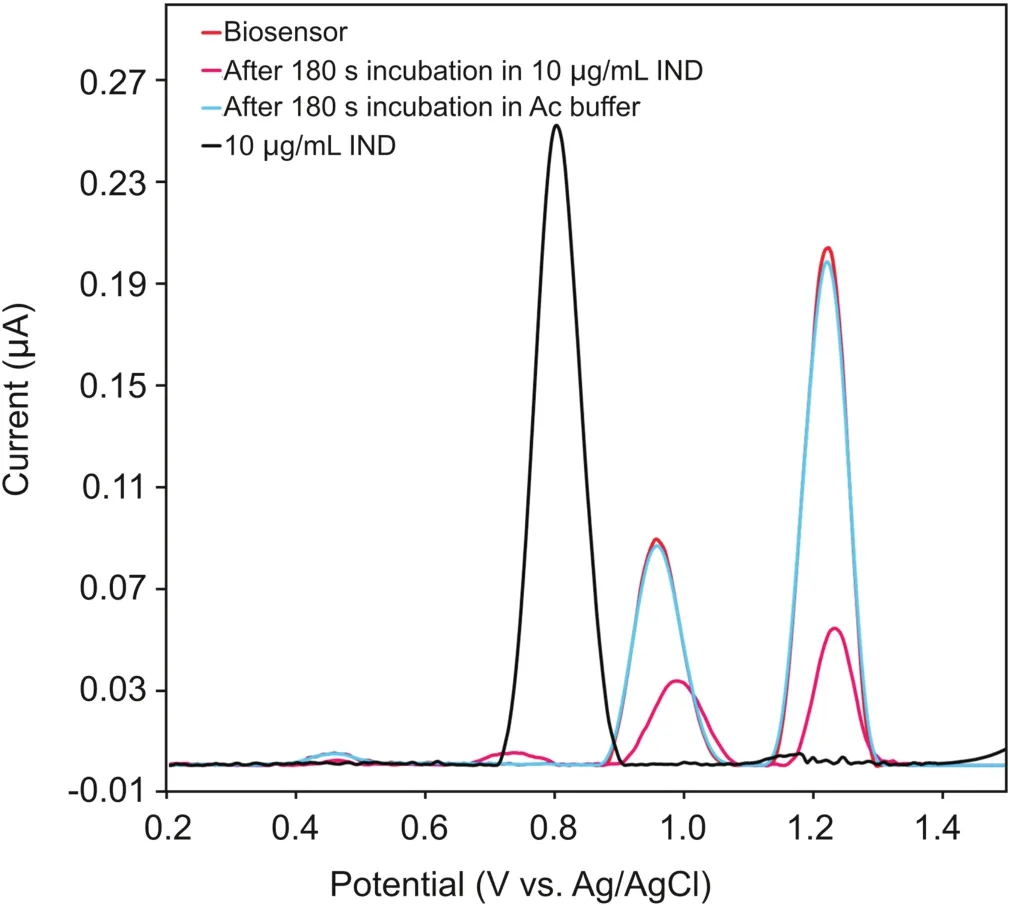
Fig.2.DP voltammograms of 10.0μg/mL IND,the ct-dsDNA biosensor,the ct-dsDNA biosensor after incubation in bare 0.1 M acetate buffer(pH=4.7),and the ct-dsDNA biosensor after incubation in 10.0μg/mL IND;Pulse amplitude= 50 mV,pulse time=70 ms,and scan rate=5 mV/s.
The interaction of IND with ct-dsDNA was investigated by DP voltammetry(DPV)in the solution phase,after different incubation times.From preliminary DPV measurements,it was observed that in the solution phase,the DPV peak current of 10μg/mL IND decreased just after mixing with 100μg/mL of ct-dsDNA(Fig.3A).Moreover,a similar decrease in the current of the dGuo peak was observed in the mixture solution of IND and ct-dsDNA compared to a solution of 100μg/mL of ct-dsDNA in the absence of IND.This decrease in the current of the IND and dGuo peaks can be explained from a reduction in the equilibrium concentration of free IND and ct-dsDNA[28].
Hence,the incubation of IND with ct-dsDNA in the solution phase was monitored by recording the DPV signal of a mixture of 10μg/mL IND and 100μg/mL of ct-dsDNA prepared in a 0.1M of acetate buffer solution(pH=4.70)at room temperature,after different incubation time periods,ranging from 0 to 240 min.The DP voltammogram of only 100μg/mL ct-dsDNA solution was recorded to control the IND-ct-dsDNA interaction.As can be seen in Fig.3B,with increasing incubation time,the current of the IND peak at approximately 0.8 V and of the dGuo peak at approximately 0.96 V gradually decreased,accompanied with a positive shift in the peak potential of IND and dGuo,which is consistent with the results from the DNA biosensor(Section 3.1.2).It should be noted that the surface of the GCE should be polished after each measurement to remove any electro-active contaminants adsorbed on its surface,which can block the electrode surface.
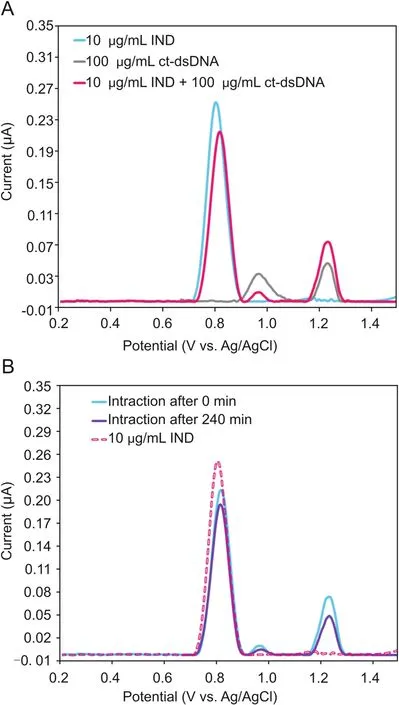
Fig.3.(A)DP voltammograms of 10μg/mL IND,100μg/mL ct-dsDNA,and of a mixture of 10.0μg/mL IND with ct-dsDNA,in the solution phase using bare GCE;(B)DP voltammograms of 10μg/mL IND and its interactions with ct-dsDNA in the solution phase,after an incubation time of 0 min or 240 min,using bare GCE.
3.1.2.In-situ study using the ct-dsDNA-electrochemical biosensor
Incubation time is an important parameter for the interaction of IND with the ct-dsDNA-coated GCE surface;hence,its optimization is necessary.In order to obtain the optimum interaction time,the oxidation signals of the ct-dsDNA biosensor were measured in the absence and presence of 10μg/mL IND after different incubation times,in the range of 30-300 s.Fig.4A shows the DP voltammograms and currents of the dGuo and dAdo peaks for ct-dsDNA as a function of interaction time with IND.As can be seen from Fig.4B,a significant decrease in the current of the dGuo and dAdo peaks was observed as the incubation time increased up to 180 s,followed by an increase in the ct-dsDNA signal after 300 s of interaction time.Therefore,an incubation time of 180 s was selected for all of the subsequent experiments.The value of the percentage of dGuo signal change(S%)is one way to interpret the interaction between ct-dsDNA and IND.This value was calculated for each incubation time based on the equation described by Carter et al.[29]:

where Ssand Sbare the dGuo oxidation signals after and before the interaction,respectively.S% values for all incubation times(30,60,120,180,and 300 s)were obtained using equation(1),and are shown in Table S2.According to the literature,if a sample has an S% value higher than 85%,it is considered non-toxic,whereas if the S% value is less 50%,it is considered toxic.When the S% value lies between 50 and 85,it is classifies as a moderately poisonous sample[29-31].
According to this theory,IND does not have any toxic effect on ct-dsDNA when the incubation time is 180 s.However,it appears to be moderately toxic following shorter incubation times.The toxicity effect of IND was decreased by increasing the interaction time from 30 s to 180 s.
The electrochemical determination of IND was performed bythe DPV technique at the ct-dsDNA/GCE biosensor.Fig.5A shows typical DP voltammograms of the ct-dsDNA-coated GCE when incubated with various IND concentrations in the range of 0.0-10.0μg/mL in the 0.1 M acetate buffer solution(pH=4.70).The current of the dGuo and dAdo peak was decreased with increasing IND concentration,which could be ascribed tothe binding of IND to ct-dsDNA on the GCE surface.This decrease in the dGuo and dAdo peak currents can be explained as possible damage of the oxidizable groups in the electroactive bases of ct-dsDNA at the GCE surface due to interactions with IND[32].The presence of a positive or negative shift in the dGuo oxidation peak potential is considered to be an indication of the binding mode,intercalation or electrostatic,respectively[28,33].As can be seen from Fig.5B,IND addition caused a positive shift in the oxidation peak potential of dGuo,suggesting that the interaction between IND and ct-dsDNA occurs through groove binding or poor intercalation.In addition,the relationship between the current of the oxidation peaks for dGuo and dAdo and the IND concentration in the range of 0.0-10.0μg/mL can be described well with the following linear equations:I(μA)= -0.0054[IND]+ 0.0842(R2= 0.9908)for dGuo,and I(μA)=-0.0145[IND]+0.2024(R2=0.9909)for dAdo.The IND LOD and limit of quantification(LOQ)were calculated using the equations LOD=3 s/m and LOQ=10 s/m,in which s is the standard deviation for measurements in the absence of IND(n=3),and m is the slope of the calibration plot.Using these equations and the calibration curves,the LOD and LOQ values for IND were found to be 0.29 and 0.98μg/mL,respectively,based on the dGuo signal,and 0.23 and 0.78μg/mL,respectively,based on the dAdo signal.Analytical figures of merits for the proposed method compared with other methods reported for this drug are given in Table 1.Repeatability and reproducibility parameters were also determined to assess the precision of the method.The repeatability of the measurements conducted with the ct-dsDNA-modified GCE was determined by measuring repeatedly the current of the oxidation peak of dGuo by DVP using the same electrode.The relative standard deviation(RSD)for the three measurements was 1.60%.The reproducibility of the measurements conducted with the ctdsDNA-modified GCE was evaluated by measuring the current of the oxidation peak of dGuo with five different electrodes.The RSD for the reproducibility study was 2.16%.These results show the excellent performance and precision of the proposed ct-dsDNA-modified GCE biosensor for the determination of IND.
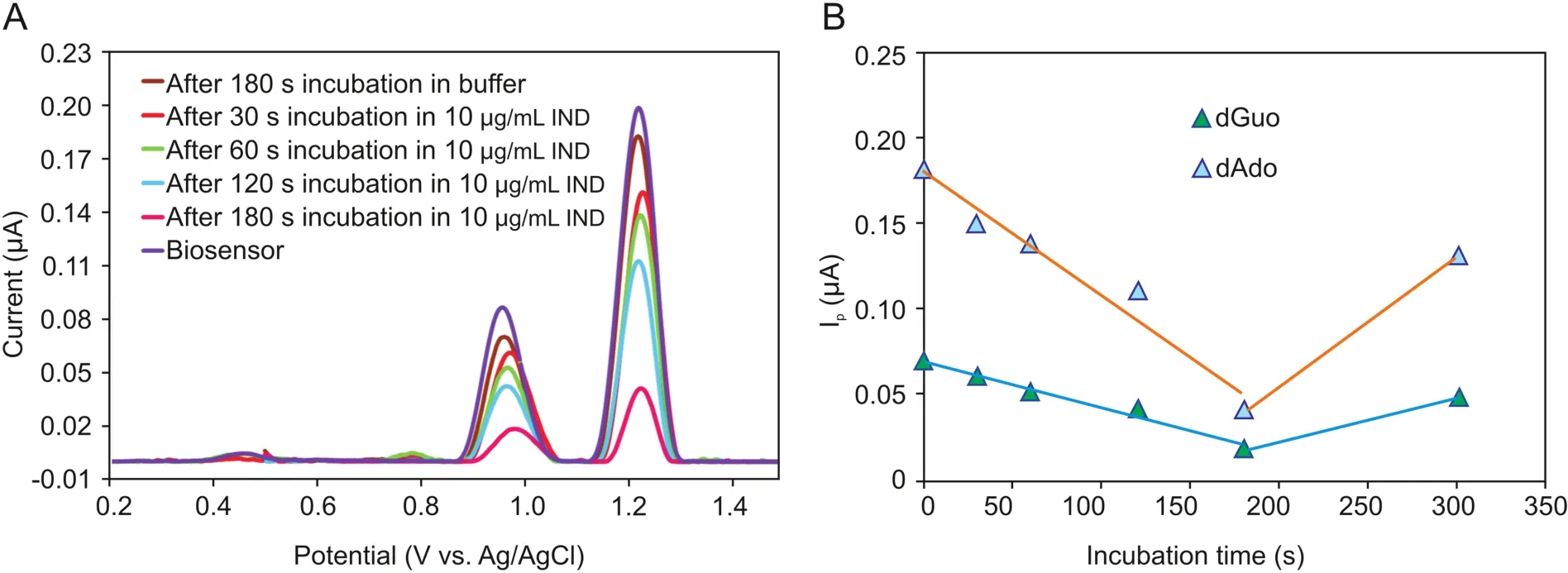
Fig.4.(A)DP voltammograms and(B)the plot of the current of the dGuo and dAdo peaks versus incubation time for the interaction of ct-dsDNA biosensor with 10μg/mL IND for different incubation times(30 s,60 s,120 s,180 s and 300 s).
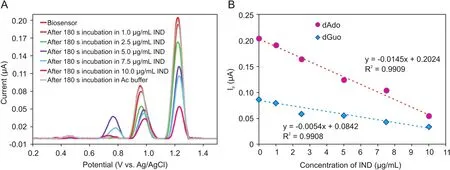
Fig.5.(A)DPV voltammograms of the ct-dsDNA biosensor after interacting with various IND concentration(1.0,2.5,5.0,7.5,and 10.0μg/mL)(incubation time:180 s),(B)Calibration plot for IND determination based on the currents of the dGuo and dAdo peaks.
3.2.Investigation of the interactions between IND and ct-dsDNA using fluorescence spectroscopy
Fluorescence titrations were conducted using the indicator displacement approach.In this competitive approach,the fluorescence signal of the indicator is first quenched by the host molecule.Upon addition of a molecule with a higher affinity toward the host than the indicator,fluorescence recovery occurs owing to the replacement of the indicator.In our study,MB was used as the indicator:MB fluorescence was quenched and red-shifted upon DNA binding (intercalative interaction)(Fig.S2A),and recovered(fluorescence ON state)when IND interacted with DNA and replaced MB(Fig.S2B).
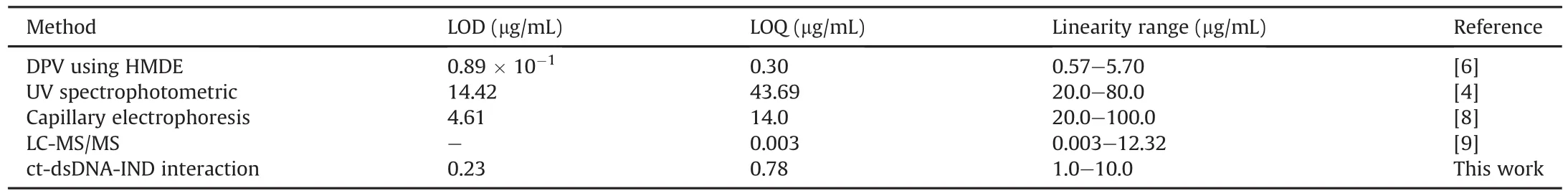
Table 1Comparison of the characteristics of the proposed method with other reported methods for quantitative determination of indinavir.
The indicator displacement assay was performed at four different temperatures to investigate the thermodynamic parameters of the IND-DNA interaction.The binding constant describing the interaction between MB and DNA was calculated using the Stern Volmer equation:

where F and F0are the fluorescence intensities in the presence and absence of DNA,respectively,τ0is the lifetime of MB fluorescence in the absence of a DNA,Kqis the MB fluorescence quenching rate constant,KSVis the Stern-Volmer constant,and[DNA]is the concentration of DNA(quencher)[34].
The slope of the plot of F0/F vs.[DNA]gives the Stern-Volmer constant,which,for the MB-DNA interactions,was at the level of~104.Following that determination,the IND titration data were evaluated using algorithms specific for the indicator displacement assay[35].The scripts implementing the algorithms were written in Origin C,and the binding constants were calculated using nonlinear regression(Fig.6).The simplest reaction describing the interactions taking place during the indicator displacement assay is:
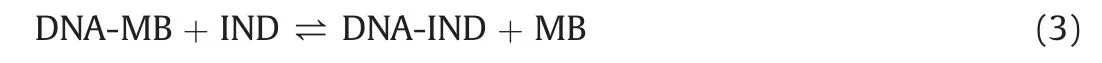
where DNA-MB is the complex between DNA and MB,DNA-IND is the complex between DNA and IND and MB represents free MB.The algorithms for the data analysis were derived based on the following binding and mass-balance equations:

where Kbis the binding constant for the interaction between IND and DNA,Kiis the binding constant for the interaction between MB and DNA,[DNA]tis the total DNA concentration,[IND]tis the total IND concentration,and[MB]tis the total MB concentration.Based on the above assumptions,the following equations were derived for the solution:
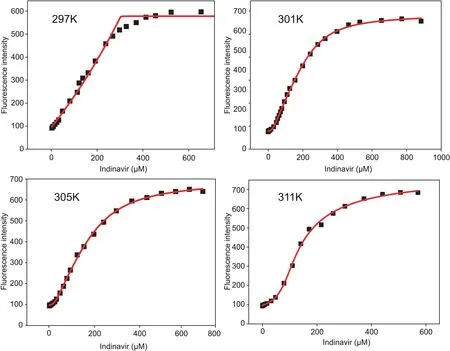
Fig.6.IND-MB+DNA titration isotherms and non-linear regression for binding constant calculations at different temperatures.
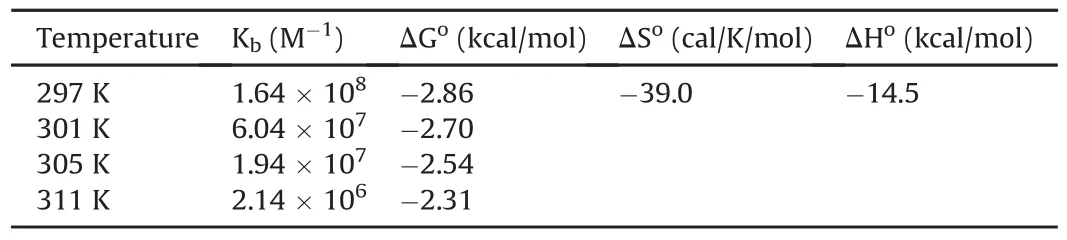
Table 2Thermodynamic parameters for indinavir-DNA interaction calculated using fluorescence titrations.
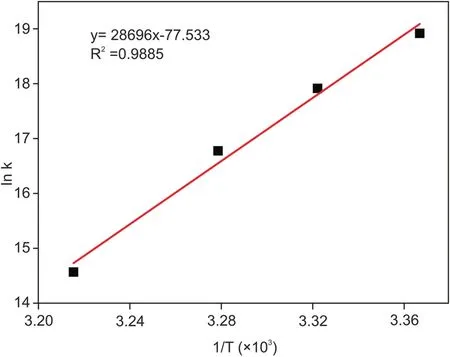
Fig.7.Van’t Hoff plot for the IND-DNA interaction.

and finally,substituting equations(11)and(12)into equation(13):
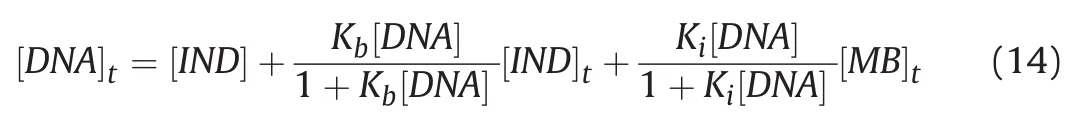
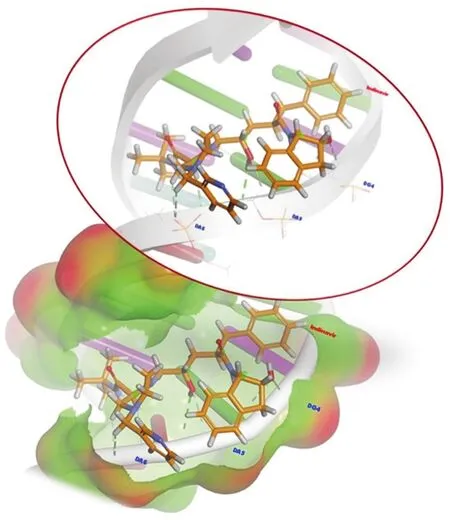
Fig.8.Global minimum energy binding conformations and interactions of IND in the binding site of DNA.
The above algorithms,which were mainly derived from Chaires et al.’s study[35],were then implemented into the relevant scripts.After defining the known values and using a set of initial estimates for the unknown parameters,the solution was computed using Newton’s method.Iterations were continued until the non-linear regression converged into a solution.
The binding constants for the IND-DNA interaction were much higher than those for the MB-DNA interaction(Table 2).Finally,binding constants computed at different temperatures were used for constructing a van’t Hoff plot(Fig.7)to derive thermodynamic parameters.Table 2 shows that the Gibbs free energy changes were negative,con firming that the interaction between IND and DNA is spontaneous.The strength of IND binding to DNA(binding constants)decreased with increasing temperature,suggesting that the process is exothermic,which is also supported by the values of enthalpy changes.
Table 3 describes the affinities and modes of binding of some drugs to DNA[20-23,36-38].Whereas other antiviral drugs,such as lamuvidine,ribavirin,and oseltamivir,show weak or moderate binding affinities,IND interacts strongly with DNA,forming eight hydrogen bonds,as revealed in our docking study.Netropsin and diminazene have higher affinities than IND,whereas daunomycin and gemicitabine have weaker binding affinities than IND.
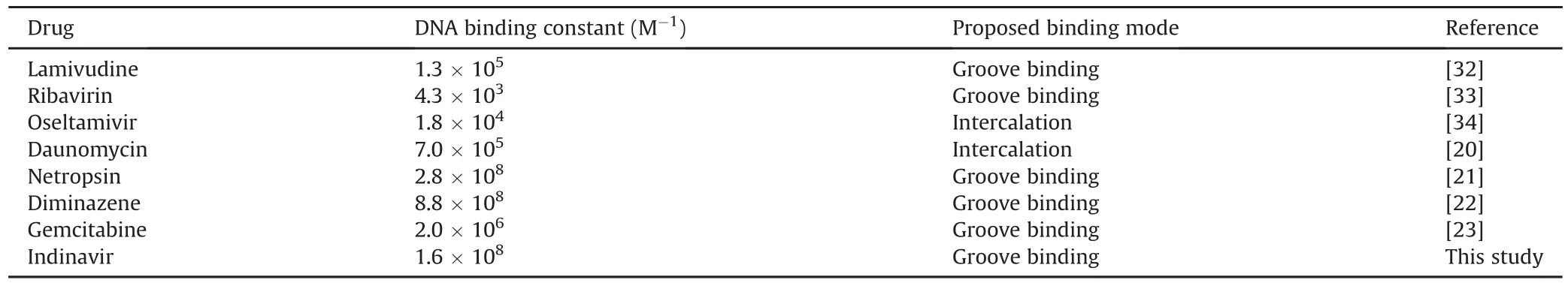
Table 3DNA binding constants of some drugs at room temperature.
3.3.Molecular modeling study of the interaction of IND with ctdsDNA
Based on the docking results,the calculated binding energyΔG(kcal/mol)and inhibition constant Ki(nM)for the interaction between IND with DNA were-8.38 kcal/mol and 726.14 nM at 298.15 K,respectively.The docking simulation generated an IND conformation inside the binding pocket of DNA with a binding energy of-8.38 kcal/mol(Fig.8).All docking solutions were examined in order to select the right conformation.Furthermore,visualization of the interaction between IND and DNA in the complex showed that all of IND structures could enter the DNA binding grooves(Fig.8).The interactions between IND and DNA were mediated by eight hydrogen bonds,the first two of which were conventional hydrogen bonds;the residues involved were carbon and hydrogen,and bonds were formed with the A:DG4,A:DA5,and A:DA6 nucleotides of the DNA molecules,as shown in Fig.8 and Table S3.
4.Conclusions
In this study,an electrochemical DNA biosensor based on ctdsDNA-modified GCE was developed for the detection of IND for the first time.The interaction of IND with ct-dsDNA was confirmed by a decrease in the dGuo and dAdo oxidation peaks.The type of interaction between IND and ct-dsDNA was examined by electrochemical,spectro fluorometric,and modeling approaches.All results suggest that the possible binding mode of IND to ct-dsDNA is based on groove binding via hydrogen bonds.The proposed electrochemical DNA biosensor is simple and sensitive for the determination of IND and possesses a low LOD.Furthermore,the developed sensing platform is a promising candidate for monitoring new dsDNA-drug interactions in pharmaceutical analysis.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
F.M.kindly acknowledges the TUBITAK for Program 2221.F.M.,B.D.T.,M.G.C.and S.A.O thanks for the support of Ankara University.IND was kindly supplied by Merck Sharp&Dohme Pharm.Ind.(Istanbul,Turkey)and we are thankful to them for their support.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.08.004.
 Journal of Pharmaceutical Analysis2020年5期
Journal of Pharmaceutical Analysis2020年5期
- Journal of Pharmaceutical Analysis的其它文章
- Selective and sensitive fluorescence imaging reveals microenvironment-dependent behavior of NO modulators in the endothelial system
- Recent progress in the molecular imaging of therapeutic monoclonal antibodies
- A carbon nanoparticle-peptide fluorescent sensor custom-made for simple and sensitive detection of trypsin
- Strategies for PET imaging of the receptor for advanced glycation endproducts(RAGE)
- Fluorescent antibiotics for real-time tracking of pathogenic bacteria
- Recent advances in construction of small molecule-based fluorophoredrug conjugates
