Molecular Cloning, Expression Pattern, and 3D Structural Prediction of the Cold Inducible RNA - Binding Protein (CIRP) in Japanese Flounder (Paralichthys olivaceus)
YANG Xiao, GAO Jinning, MA Liman, LI Zan, WANG Wenji, WANG Zhongkai,YU Haiyang, QI Jie, WANG Xubo, WANG Zhigang, and ZHANG Quanqi
?
Molecular Cloning, Expression Pattern, and 3D Structural Prediction of the Cold Inducible RNA-Binding Protein (CIRP) in Japanese Flounder ()
YANG Xiao, GAO Jinning, MA Liman, LI Zan, WANG Wenji, WANG Zhongkai,YU Haiyang, QI Jie, WANG Xubo, WANG Zhigang, and ZHANG Quanqi*
,,,266003,..
Cold-inducible RNA-binding protein (CIRP) is a kind of RNA binding proteins that plays important roles in many physiological processes. Thehas been widely studied in mammals and amphibians since it was first cloned from mammals. On the contrary, there are little reports in teleosts. In this study, the Pogene of the Japanese flounder was cloned and sequenced. The genomic sequence consists of seven exons and six introns. The putative PoCIRP protein of flounder was 198 amino acid residues long containing the RNA recognition motif (RRM). Phylogenetic analysis showed that the flounder PoCIRP is highly conserved with other teleost CIRPs. The 5’ flanking sequence was cloned by genome walking and many transcription factor binding sites were identified. There is a CpGs region located in promoter and exon I region and the methylation state is low. Quantitative real-time PCR analysis uncovered that Pogene was widely expressed in adult tissues with the highest expression level in the ovary. The mRNA of the Powas maternally deposited and the expression level of the gene was regulated up during the gastrula and neurula stages. In order to gain the information how the protein interacts with mRNA, we performed the modeling of the 3D structure of the flounder PoCIRP. The results showed a cleft existing the surface of the molecular. Taken together, the results indicate that the CIRP is a multifunctional molecular in teleosts and the findings about the structure provide valuable information for understanding the basis of this protein’s function.
cold-inducible RNA-binding protein; CIRP;; expression pattern; 3D modeling; CpGs
1 Introduction
Cold-inducible RNA-binding protein (CIRP, also named CIRBP) was first cloned from mammals as a cold-shock protein by Nishiyama. ( 1997). The CIRP is one of the highly conserved glycine-rich RNA-binding proteins which contain an amino-terminal domain named consensus sequence of RNA-binding domain (CS-RBD) or RNA recognition motif (RRM) and a carboxyl-terminal glycine-rich domain (GRD) (Bergeron, 1993).
CIRP is not only induced in response to mild hypothermia(Nishiyama, 1998a; Tong, 2013) but also involved in many other biological processes (Liu, 2013; Nishiyama., 1997). It could be induced by DNA damage through UV irradiation (Yang & Carrier, 2001), osmotic stress(Pan, 2004), low oxygen level (Wellmann, 2004), light (Sugimoto and Jiang, 2008), diseases (cancers and ischemia) (Hamid, 2003, Zhou, 2009) and damages (Xue, 1999){Yang, 2001 #14730}. In ad-dition to these stress induction, CIRP is expressed in brain associated with hibernation (Saito, 2000) and may play important roles in pronephrogenesis and neurogenesis during the embryonic period of(Uochi and Asashima, 1998). Recent study also found that it may modulate circadian gene expression., 2013, Morf, 2012) and is associated with circadian rhythm (Morf., 2012; Nishiyama, 1998b;Ripperger, 2000). In the mouse testis the expression ofduring spermatogenesis and after heat stress has special effects in the primary spermatocytes through regulating target mRNA stabilization (Nishiyama., 1998a; Xia, 2012). In, however, CIRP was found to be the major cytoplasmic RNA-bind- ing protein (Matsumoto, 2000). This may suggests that the CIRP may have different roles between the males and females.
Although CIRPs have been explored in many species including mammals and amphibians, teleost CIRP and the methylation level of this gene have not been reported in detail. The CIRP has been reported to be induced by osmotic stress in salmon (Pan., 2004). Screening of EST-SSRs markers revealed that CIRP is related to growth traits of common carp (Ying, 2009). A cDNA-AFLP analysis identified the CIRP gene as one of female specific expressed sex-related genes in flatfish (Taboada, 2012). As is known that female flatfish grows faster and is bigger than the males, the related reports implied that the CIRP may be involved in this phenomenon. In order to better understand its biology functions in fishes and its relationship with sex, we cloned the full-length cDNA, the genomic sequence and the 5’ promoter region of CIRP gene from Japanese flounder(Po), a commercially important marine fish for both fisheries and aquaculture in East Asian countries. We quantified its expression in different adult tissues and at several early developmental stages to get knowledge about its possible functions during embryogenesis as well as adult growth. We also analyzed the potential regulatory motif and the methylation level of CpGs located in the promoter region. Phylogenetic reconstruction and 3D structural prediction were also applied to provide more information.
2 Materials and Methods
2.1 Animals and Sampling
The fishes were sampled from Haiyang Yellow Sea Aquatic Product Co., Ltd, Yantai, Shandong, China. Six adults (three females and three males) were randomly selected from the tank. Eight tissues (heart, liver, spleen, kidney, whole bran, intestine, ovary, testis and skeletal muscle) were dissected and snapped frozen in liquid nitrogen and then kept at ?80℃ before using. Artificially fertilized eggs were incubated at 15℃ in tank with continuous aeration. Embryos of different developmental stages (including unfertilized eggs, one cell period, eight cells period, multi cells stages, high blastula, gastrula, neurula, tail-bud stage, heart-beating stage and hatching stage) were collected and directly preserved in 1mL RNAwait liquid (Solarbio, SR0020, Shanghai) and then kept at ?80℃ before use. A small piece of muscle tissue was saved at ?80℃ for genomic DNA extraction.
2.2 Extraction of Genomic DNA and RNA
Genomic DNA was extracted from muscle by phenol chloroform extracting method. Briefly, the tissues grinded in liquid nitrogen were added to 600μL TNES (125mmolL?1NaCl, 10mmolL?1Tris-HCl, 10mmolL?1EDTA, SDS 0.5%, Urea 4molL?1, pH7.4) buffer. Then the samples were digested overnight at 56℃ by 6μL proteinase K (20mg mL?1) at the final concentration of 200μgmL?1. The DNA was then isolated with phenol and chloroform/isoamyl alcohol, precipitated with isopropanol and washed with 70% ethanol. The isolated DNA were then dissolved in ddH2O, degraded with RNaseA at 37℃ for 1h, and then preserved at ?20℃ until use.
Total RNAs were extracted from adult tissues and embryos of different developmental stages using Trizol reagent (Invitrogen) according to the manufacturer’s instruction and kept at ?80℃before use.
2.3 cDNA Cloning and Sequence Analysis ofPo
Total RNA (1μg) from the ovary tissue was reverse transcribed to synthesize the first strand cDNA using MMLV-Reverse-Transcriptase (RNase H?) (TaKaRa, Dalian, China). Primers for cloning the initial fragment of PomRNA were designed according to conserved sequences predicted in other teleosts (Table 1). The PCR program was as follows: 94℃ pre-denaturation for 10min; 30cycles of 94℃ denaturation for 30s, 51℃ annealing for 30s, 72℃ extension for 30s; and then final elongation at 72 for 10min.
In order to get the full length of the cDNA sequence,gene specific primers for 5’ and 3’ RACE were designed according to the obtained sequence fragment (Table 1). We conducted the experiments to obtain the 5’ and 3’ unknown cDNA regions by using the SMART RACE PCR Amplification Kit (Clontech, CA , USA) following the manufacture’s protocol. The RACE PCR conditions were: 94℃ for 5min; 24 cycles of 94℃ for 30s, 58℃ for 30s, 72℃ for 3min; and a final elongation at 72℃ for 10min. The full-length of the coding sequence was amplified with two specifically designed primers (Table 1) using the following program: 94℃ for 5min; 30 cycles of 94℃ for 30s, 55℃ for 30s, 72℃ for 3min; and a final elongation at 72℃ for 10min.

Table 1 Primers used in this study
All the PCR reactions were performed using a GeneAmp PCR system 9700 (Applied Biosystems, Forster City, CA, USA), and the amplification products were separated by 1% agarose gel electrophoresis, purified by using the Gel DNA recovery Kit (Zymoclean, CA, USA), cloned to pMD18-T vector (TaKaRa, Dalian, China), and transformed to competentTrans5α strain (TransGenBiotech, Beijing, China). Positive clones containing the plasmid with inserts of the appropriate sizes were identified by colony PCR using M13Fw and M13Rv primers and sequenced.
The ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/ gorf.html) was used to predict the open reading frame (ORF).
2.4 Analysis of Gene Sequence and Promoter of Po
In order to know the genomic structure of Powe amplified the genomic DNA fragment to get the introns with primers as showed in Table 1. The PCR cycles were described as follows:94℃ for 5min; 30 cycles of 94℃ for 30s, 55℃ for 30s, 72℃ for 3min; and a final elongation at 72℃ for 10min. The promoter region was amplified by using Genome Walking Kit (TaKaRa, Dalian, China) with gene specific primers listed in Table 1. The PCR program was designed following the instructions of the Kit.
The gene structure of the exon-intron was estimated by the Spidey program in NCBI homepage (http://ncbi.nlm. nih.gov/IEB/Research/Ostell/Spidey/). The putative transcription start site (TSS), TATA box and CAAT box were predicted by using the tools of Promoter 2.0 Prediction Server (http://www.cbs.dtu.dk/services/Promoter/) and SIGSCAN (http://www-bimas.cit.nih.gov/molbio/signal/). Alibaba v2.1 (http://www.gene-regulation.com/pub/programs/alibaba2/index.html) was used to predict the transcription factor (TF) binding sites. The CpGs island was predicted by EMBOSS Cpgplot (http://www.ebi.ac.uk/ Tools/seqstats/emboss_cpgplot/).
2.5 Methylation Level of the CpGs in Promoter and Exon 1 Region
The DNA samples of six tissues were extracted from 3 females and 3 males respectively using the method mentioned above. The samples were treated with the EZ DNA Methylation-GoldTMKit following the manual instruction, and used for PCR with the primers listed in Table 1. The amplicons were purified, cloned and sequenced as described above. For each sample, 10 positive clones were sequenced and the results were analyzed with BDPC (Bisulfite sequencing Data Presentation and Compilation) (http://services.ibc.uni-stuttgart.de/BDPC/).
2.6 Quantitative Real-Time PCR (qPCR) of Po
Total RNA extracted from different tissues of six healthy adults (three females and three males) and different developmental stages was used to analyze the expression pattern using qPCR. The embryos or unfertilized eggs at different development stages were pooled for qPCR with each pool containing 20 eggs or embryos. Three pools were prepared for each stage as triplicates. The cDNA were generated from RNA samples through the method described above. The primers (Table 1) used for qPCR were designed with Primer Express v3.0 (Applied Biosystems). The amplification conditions were performed according to the manual book of SYBR Premix?EX TaqTMII (TaKaRa, Bio Inc., Shiga, Japan). 18S rRNA was selected as the reference gene (Zhong, 2008) and used to normalize the mRNA levels of target genes in all samples. Negative control was performed at the same time but no template was added. The amplicfication was performed in an ABI Prism 7500 Sequence Detection System (Applied Biosystems, Forster, CA, USA). The program was as follows: 1 cycle of 95℃ for 2min; 40 cycles of 95℃ for 15s, 60℃ for 45s; 1cycle of 95℃ 15s, 60℃ 1min, 95℃ 15s, 60℃ 15s.
We constructed standard curve with a serial dilution of solution which contained certain amount of plasmids inserted with either 18Sor the Pogene fragments. Experiment with each sample was conducted in triplicate and the means were used for further analyses using the relative standard curve.
2.7 Sequence Alignment and Phylogenetic Reconstruction
We collected homologous sequence of other species’ CIRPs, and made the multiple alignments of the CIRPs by ClustalX 2.1software (Larkin, 2007). The phylogenetic reconstruction and molecular evolutionary analyses were conducted by using the MEGA version 5 (Tamura, 2011). And the tree was constructed using the neighbor joining method with 1000 bootstrap replicates.
2.8 Analysis of the PoProtein Sequence and Modeling of Its 3D Structure
The protein sequence was analyzed for its molecular mass and theoretical isoelectric point using the Compute PI/Mw tool (http://web.expasy.org/compute_pi/). The P- fam (http://pfam.sanger.ac.uk/) databases were used to scan the domains in the protein sequence.
Three-dimension structure of the Japanese flounder Poprotein was predicted using SWISS-MODEL server (http://www.expasy.org/swissmod/SWISS-MODEL. html) and the PDBsum Generate (http://www.ebi.ac.uk/ thornton-srv/ databases/ pdbsum/Generate.html) was used to predict the molecular surface and cleft in the molecular.
2.9 Statistical Analysis
The expression results were analyzed with SPSS19.0 software. Data reported were logarithmically transformed before statistical analysis. Significant differences between samples were determined by one-way or two-way ANO- VA.multiple comparisons were carried with LSD multiple range tests. Data obtained from between samples were considered significantly different when< 0.05. The data are displayed using the mean±standard error of the mean.
3 Results and Discussions
3.1 The Cloning of the mRNA Sequence of the Japanese Flounder(Po)
A 597bp long cDNA fragment of the Pogene was cloned from ovary with the primers cirp-m-fw/rv. The unknown 5’ and 3’ regions of the cDNA fragments were obtained through the RACE reaction with the primers RACE-5-rv and RACE-3-fw, respectively. The full length of the cDNA sequence was obtained by assembling all the sequences according to the overlaps between the fragments and verified using the gene specific primers cirp-w-fw/rv. The mRNA sequence of the Pois of 1596bp with an 85bp 5’ UTR (untranslated region), a 914bp 3’UTR and a 597bp coding sequence (CDS) predicted by ORF Finder (Fig.1). A non-canonical polyade-nylation signal site (ATTAAA) was found in the 3’ UTR near the polyA site.

Fig.1 The mRNA sequence of PoCIRP. CDS is shown in capital letters, whereas the 5’UTR and 3’UTR are shown in small letters. The deduced amino acid sequence is shown by single letter code of amino acids below the CDS. The non-canonical polyadenylation signal site (ATTAAA) is boxed.
The predicted amino acid sequence of the PoCIRP consists of 198 amino acid residues with the calculated molecular mass of 20.6kDa. The PoCIRP was predicted to contain the RNA recognition motif (RRM_1, PF00076) from the 7th to 77th residues and several Arg-Gly-Gly (RGG) repeats in the glycine-rich region, which is the same as reported in salmon, human and mice (De Leeuw, 2007; Jin, 2009). BLASTP analysis showed that the PoCIRP was similar to other CIRPs of mammals, amphibians and teleosts. The sequence identities were 47%, 75%, 73%, 69%, compared to,,, respectively. The genomic, mRNA sequence and the predicted protein sequence were submitted to NCBI and the accession NO. is KJ739525.
3.2 Phylogenetic Analysis of Poin Vertebrates
In order to understand the relationships of PoCIRP among the vertebrate CIRPs, we constructed the phylogenetic tree with CIRPs of 16 sequences using human RBM3, a member of highly conserved family of RNA binding proteins, as outgroup (Fig.2). The CIRPs were divided into four clusters which agree with taxonomic status of these species, and the PoCIRP was located with- in the teleosts group with 76% bootstrap support, indicating that this protein is one of the fish CIRP orthologs and these proteins are highly conserved proteins.

Fig.2 The phylogenetic tree based on amino sequence alignment for CIRPs in vertebrates. Bootstrap values (%) indicate 1000 replicates. Homo sapiens RBM3 was used as outgroup. The number of each internal branch is the local bootstrap probability. The GenBank accession number of the proteins are as follows: Homo sapiens (BAA11212.1), Pongo abelii (NP_001124692.1), Sus scrofa (NP_001233126.1), Rattus norvegicus (BAA19092.1), Mus musculus (BAA11213.1), Cricetulus griseus (NP_001231002.1), Gallus gallus (AGS43093.1), Xenopus laevis (A: NP_001079794.1, B: NP_ 001080069.1), Xenopus (Silurana) tropicalis (NP_001017228.1), Oncorhynchus mykiss (ACO08387.1), Salmo salar (ACI 33459.1), Danio rerio (AAI15178.1), Paralichthys olivaceus (KJ739525), Anoplopoma fimbria (ACQ58801.1), Osmerus mordax (ACO09027.1), RBM3 human (P98179.1).
3.3 Identification and Analysis of the Genomic Structure of the Po
Whole genome sequence of Powas obtained with specific primers (Table 1). The introns were identified by comparing the mRNA sequence with the gene fragment (Fig.3). The Pogene had six introns which were 661 bp, 110bp, 161bp, 95bp, 94bp and 516bp, respectively. All the introns were found to be spliced conforming to the GT-AT rule. The 5’ UTR was encoded by the exon I and a part of the exon II, the 3’ UTR was encoded by most of the exon VII and the CDS was encoded by the rest parts of the exons.
Nearly about 4000bp of the upstream region of Powas sequenced by genome walking method. The putative transcription start site (TSS), TATA box and CAAT box predicted by Promoter 2.0 Prediction Server and SIGSCAN were shown in Fig.4. There was a high likely prediction of the TSS site upstream, the sequenced start site of the mRNA. As there was the possibility that the mRNA molecular was not complete when it was extracted, the predicted TSS may be right. Many transcription factor binding sites were predicted, such as Oct-1, AP-1, SP1, C/EBP,. (Fig.4). The regulation regions of the Pogene may be conserved as it has been reported that there are some AP-1 binding sites in enhancer of the genes (Sumitomo, 2012). Many HSF and several SRY binding sites could be found in the promoter region. The existence of these HSF binding sites implied that the gene express level of Pocould be regulated by the temperature shift. In our previous study we also found many HSF and SRY binding sites in the promoter region of heat shock protein (HSP) 70KDa (Qi., 2014). Although the genetic sex determination of the Japanese flounder was reported to be XX/XY type, it could easily be influenced by high temperature (Yama- moto, 1999). Temperature shift might change the expression levels of CIRP and HSP through the HSF. The existence of several SRY binding sites in the promoter region might indicate that these genes may also influence/or be associated with gonad development and sex differentiation.

Fig.3 The genomic structure of the PoCIRP gene. The exons are shown as boxes, the introns are represented with black lines and the upstream region is represented with gray line. The CDS regions are shown with black boxes and the UTR regions are shown with white boxes.

Fig.4 The predicted TSS, TATA box, CpGs island and transcription factor binding sites. The binding sites of different transcription factors are marked with different symbols. The vertical bars on the second line are used to represent the CpGs sites.
3.4 The mRNA Expression Pattern of Poin Adult Tissues
We performed the experiments to study the expression level of PomRNA in different flounder tissues, and the RT-qPCR was designed using the same amounts of RNAs from different tissues. The result revealed that PomRNA was expressed in all the tissues tested. The highest expression was detected in gonads, especially high expression was found in the ovary. The difference between testis and ovary was significant (<0.05). In all other tissues the Poexpression levels were not very high (Fig.5). Similar result has been reported in turbot, in which the predictedEST is specifically expressed in females (Taboada., 2012). Inwas reported to be found specifically in oocytes (Matsumoto., 2000). Our results, together with the previously re- ported ones, indicate that CIRP may be important for the physiological functions of ovary development or may have special functions in oogenesis and oocyte maturation.

Fig.5 The quantitative analysis of PoCIRP expression in adult tissues. The relative expression variance is shown as ration (the amounts of PoCIRP were normalized to the corresponding 18S rRNA values).The data are shown as mean ± SD (n=3). Two way ANOVA analysis was performed to detect the significant differences between means. Columns with different letters show significant difference (P<0.05).
The existence ofmRNA in many tissues of flounder such as brain and heart suggests that this gene might play many other roles in this fish. CIRP may play roles of protection for testis, brain and heart as reported in earlier papers (Liu, 2010; Xia., 2012).
3.5 Methylation Analysis of the CpGs in the Pro-moter and Exon 1 Region
The 229nt putative CpGs islands located in the region of exon I were predicted using EMBOSS Cpgplot. The relative positions between the predicted CpGs and the TF binding sites are showed in Fig.4. It happened that some of the CpG sites were adjacent to the TF binding site, such as C/EBP at 3567bp, EmBP-1 at 3584bp, myogenin at 3596bp and Sp1 at 3639bp (Fig.4). This indicates that the methylation of these sites may have influence on the expression of the CIRP gene. The promoter and exon I region’s GpGs methylation levels range from 0.0%-1.2% in different tissues of male and female (Fig.6).The detected methylation CpGs sites are located in the sites of 7th, 8th, 9th, 10th, 11st, 12nd, 14th, 15th CpG site. The detected methylation sites were related to the C/EBP, REB1, Sp-1 and EmBP-1. We compared the methylation level between the two factors: sexes and various tissues based on two way ANOVA analysis. The-value was 0.425 and the-value was 0.827 between male and female, which indicates that there is no significant difference between the sexes in methylation level. Meanwhile, the methylation level among different tissues also showed no significant difference as the-value was 0.048 and the-value was 0.828. It is known that the DNA methylation status can influence the gene expression level (Tate and Bird, 1993). So the results indicate that the Pomight be widely expressed in males’ and females’ different tissues and this has been proved in 3.4. And the expression level of the Pois not mainly regulated by the methylation status of the CpGs site.

Fig.6 The methylation levels of the CpGs sites in the promoter region of both sex and different tissues. Ten clones were analyzed per fish. Result is showed as mean ± SE. The Y-axis value is the percentage of the methylated sites. Groups with different letters are significantly different (P<0.05). The methylation levels of all the groups show no significant difference.
3.6 The Expression Pattern of PoDuring Developmental Stages
The qPCR analysis showed a low level of maternal deposit ofmRNA in the fertilized eggs of Japanese flounder. The mRNA level was kept steady until multicell stages, began to increase at blastula stage and increased sharply at gastrula stage (Fig.7). The Poexpression reached the highest level at the neurula stage, and decreased afterward reaching relatively lower levels from tail-bud stage to hatching (Fig.7). One-way ANOVA analysis showed significant variations between different stages (<0.05).
It has been reported that CIRP participates in many processes during the developmental stages (Peng, 2006; Tanaka, 2000; van Venrooy, 2008).was found to be expressed transiently in developing pronephros and neural tissues (Uochi and Asashima, 1998). However, there has been no direct experimental evidence showing the developmental regulation ofexpression in teleost fish. In the present study, the highest PomRNA level was observed at neurula stages. This is in agreement with the result obtained in, which may suggest its possible function in the development of the neural system in amphibians and teleosts. It is also noticeable that the embryonic expression, or the increase of expression, of Pobegan at blastula stage. This timing of increase is earlier than the differentiation of germ layers, but is the same as the expression of some genes specific for embryonic stem cells (ESC) and/or primordial germ cells (PGC) in Japanese flounder (Gao, 2013; Wu, 2014). Therefore, whether or not the start of Poexpression is associated with the formation of primitive cells for different germ layers, or with the appearance of ESC and PGC, will be of great interest.
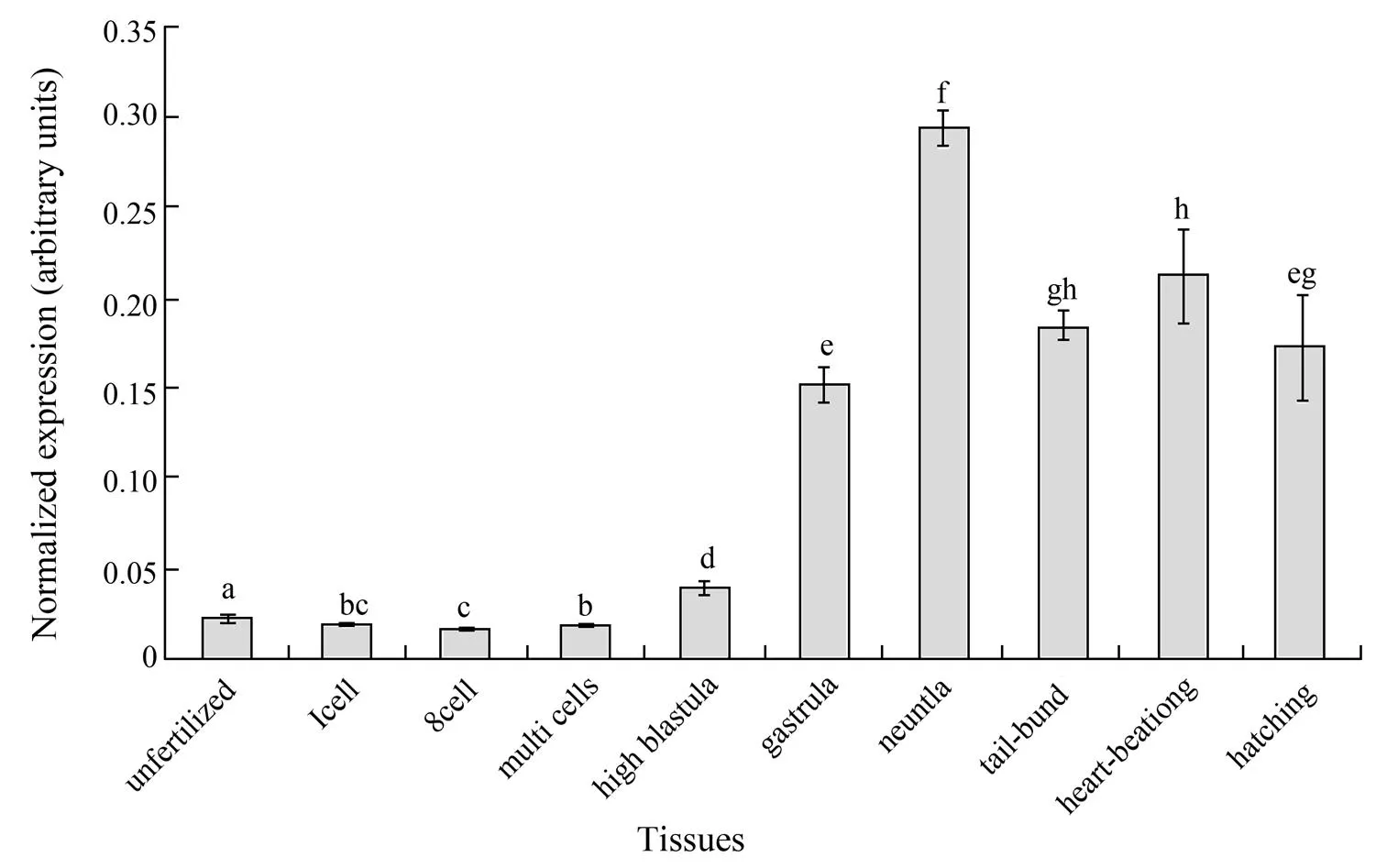
Fig.7 The quantitative analyses of PoCIRP expression during the developmental stages. The relative expression variance was shown as ration (the amounts of PoCIRP were normalized to the corresponding 18S rRNA values). The data was shown as mean±SD (n=3). One way ANOVA analysis was performed to detect the significant differences between means. Columns with different letters show significant difference (P<0.05).
3.7 3D Modeling of CIRP
The 3D model of PoCIRP was predicted with the Swiss-Model Workspace (Fig.8A). The solution stucture of RRM domain (1×5sA) was selected as the template model (Fig.8B) and the sequence identity was 69.51% between PoCIRP and the model sequence. The modelled residue sites ranged from 2 to 83, containing the RNA recognition motif. For assessment part, QMEANscore4 was 0.78, showing high quality of the model, QMEAN Z-score was ?0.17. All the assessment indicated that the 3D structure of PoCIRP model was of a good quality. When superposed to the template structure, the model showed very good overlapping (Fig.8C). By comparing the two models, we found that the target protein had similar composition to the second structures, such as helices and beta-strands, and that the spatial structure and location of these compositions were nearly the same. This also indicates that CIRPs are highly conserved proteins. With PDBsum Generate tools, the potential RNA binding site and the molecular surface cleft of PoCIRP were successfully displayed. There was a cleft between the binding domains (Fig.8D). This predicted structure may explain that the PoCIRP (RNA binding protein) interacts with mRNAs though the cleft.

Fig.8 A, Model of the PoCIRP predicted by swiss program; B, The 3D structure of the template sequence(1×5sA) in the RCSB database; C, A structure superposition of the PoCIRP and the template. The yellow band stands for the template sequence and the green band stands for the target sequence; D, The molecular surface and cleft of the PoCIRP predicted by PDBsum generate.
4 Conclusion
In this study, we identified the mRNA and genomic sequences of Pofromwhich is a highly conserved protein in teleosts. Many TF binding sites were predicted, especially the HSF binding sites and SRY binding sites in the enhncer and promoter region of the Pogene. This could explain two phenomena. The first is that the expression level of thegoes down when temperature rises (Nishiyama., 1998a) and the second is that the temperature has influence on the sex ratio of Japanese flounder. In addition, a putative CpGs island was predicted and low methylation level was identified. The qPCR results verified a wide expression in all tissues with the highest expression level existing in ovary. Expression profiling during development stages indicated that the CIRP might be involved in the development of the neural system. The 3D structure prediction also gave information about the mechanism of the interaction between CIRP and mRNAs. The present study provides the basic information about the Poof the Japanese flounder and will benefit further studies on the function of this gene.
Acknowledgements
The work was supported by the National High Technology R&D Program of China (2012AA10A402) and the National Natural Science Foundation of China (31172385).
Bergeron, D., Beauseigle, D., and Bellemare, G., 1993. Sequence and expression of a gene encoding a protein with RNA-binding and glycine-rich domains in., 1216 (1): 123-125.
De Leeuw, F., Zhang, T., Wauquier, C., Huez, G., Kruys, V., and Gueydan, C., 2007. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor., 313 (20): 4130-4144, DOI: 10.1016/j.yexcr.2007.09.017.
Gao, J., Wang, J., Jiang, J., Fan, L., Wang, W., Liu, J., Zhang, Q., and Wang, X., 2013. Identification and characterization of a nanog homolog in Japanese flounder ()., 531 (2): 411-421, DOI: 10.1016/j.gene.2013.08.030.
Gu, Y., Cao, D. C., Zhang, Y., Chang, Y., Li, O., Zhang, X., Lu, C., and Sun, X., 2009. Screening of EST-SSRs markers related to growth traits in common carp (L.)., (1): 15- 22 (in Chinese with English abstract).
Hamid, A. A., Mandai, M., Fujita, J., Nanbu, K., Kariya, M., Kusakari, T., Fukuhara, K., and Fujii, S., 2003. Expression of cold-inducible RNA-binding protein in the normal endo- metrium, endometrial hyperplasia, and endometrial carcino- ma., 22 (3): 240-247, DOI: 10.1097/01.pgp.0000070851.25718.ec.
Jin, F., Pan, Y., Li, S., Yang, H., Ji, H., Zhao, Q., and Yin, W., 2009. Cloning and sequence analysis of cold inducible RNA-binding protein cDNA from testis tissue in BALB/C Mice., 15 (1): 87-90, DOI: 10.3724/SP.J.1145.2009.00087.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G., 2007. Clustal W and clustal X version 2.0., 23 (21): 2947-2948, DOI: 10.1093/bioinfor- matics/btm404.
Li, S. Z., Jin, F. H., and Pang, Y., 2009. Cloning and Sequence Analysis of Cold Inducible RNA-binding Protein cDNA from Testis Tissue in BALB/C Mice., 15 (1): 87-90.
Liu, A. J., Zhang, Z. W., Li, A. M., and Xue, J. H., 2010. Effects of hypothermia and cerebral ischemia on cold- inducible RNA-binding protein mRNA expression in rat brain., 13 (47): 104-110, DOI: 10.1016/j.brainres. 2010.05.029.
Liu, Y., Hu, W., Murakawa, Y., Yin, J., Wang, G., Landthaler, M., and Yan, J., 2013. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation., (3): 2054, DOI: 10.1038/ srep02054.
Matsumoto, K., Aoki, K., Dohmae, N., Takio, K., and Tsujimoto, M., 2000. CIRP2, a major cytoplasmic RNA-binding protein inoocytes., 28 (23): 4689- 4697, DOI: 10.1093/nar/28.23.4689.
Morf, J., Rey, G., Schneider, K., Stratmann, M., Fujita, J., Naef, F., and Schibler, U., 2012. Cold-inducible RNA-binding protein modulates circadian gene expression posttran- scrip- tionally., 338 (6105): 379-383, DOI: 10.1126/ science.1217726.
Nishiyama, H., Danno, S., Kaneko, Y., Itoh, K., Yokoi, H., Fukumoto, M., Okuno, H., Millan, J. L., Matsuda, T., Yoshida, O., and Fujita, J., 1998a. Decreased expression of cold-inducible RNA-binding protein (CIRP) in male germ cells at elevated temperature., 152 (1): 289-296.
Nishiyama, H., Itoh, K., Kaneko, Y., Kishishita, M., Yoshida, O., and Fujita, J., 1997. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth., 137 (4): 899-908.
Nishiyama, H., Xue, J. H., Sato, T., Fukuyama, H., Mizuno, N., Houtani, T., Sugimoto, T., and Fujita, J., 1998b. Diurnal change of the cold-inducible RNA-binding protein (CIRP) expression in mouse brain., 245 (2): 534-538, DOI: 10.1006/ bbrc.1998.8482.
Pan, F., Zarate, J., Choudhury, A., Rupprecht, R., and Bradley, T. M., 2004. Osmotic stress of salmon stimulates upregulation of a cold inducible RNA binding protein (CIRP) similar to that of mammals and amphibians., 86 (7): 451-461, DOI: 10.1016/j.biochi.2004.06.006.
Peng, Y., Yang, P. H., Tanner, J. A., Huang, J. D., Li, M., Lee, H. F., Xu, R. H., Kung, H. F., and Lin, M. C. M., 2006. Cold-inducible RNA binding protein is required for the expression of adhesion molecules and embryonic cell movement in., 344 (1): 416-424, DOI: 10.1016/ j.bbrc.2006.03.086.
Qi, J., Liu, X., Liu, J., Yu, H., Wang, W., Wang, Z., Zhang, Q., 2014. Molecular characterization of heat shock protein 70 (HSP 70) promoter in Japanese flounder (), and the association of Pohsp70 SNPs with heat-resistant trait., 39 (2): 503-511, DOI: 10.1016/j.fsi.2014.05.038.
Ripperger, J. A., Shearman, L. P., Reppert, S. M., and Schibler, U., 2000. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP., 14 (6): 679-689, DOI: 10.1101/gad. 14.6.679.
Saito, T., Sugimoto, K., Adachi, Y., Wu, Q., and Mori, K. J., 2000. Cloning and characterization of amphibian cold inducible RNA-binding protein.-, 125 (2): 237-245, DOI: 10.1016/s0305-0491(99)00174-1.
Sugimoto, K., and Jiang, H., 2008. Cold stress and light signals induce the expression of cold-inducible RNA binding protein (CIRP) in the brain and eye of the Japanese treefrog ()..,, 151 (4): 628-636, DOI: 10.1016/j.cbpa.2008.07.027.
Sumitomo, Y., Higashitsuji, H., Liu, Y., Fujita, T., Sakurai, T., Candeias, M. M., Itoh, K., Chiba, T., and Fujita, J., 2012. Identification of a novel enhancer that binds Sp1 and contributes to induction of cold-inducible RNA-binding protein (CIRP) expression in mammalian cells., 12 (1): 72, DOI: 10.1186/1472-6750-12-72.
Taboada, X., Robledo, D., Del Palacio, L., Rodeiro, A., Felip, A., Martinez, P., and Vinas, A., 2012. Comparative expression analysis in mature gonads, liver and brain of turbot () by cDNA-AFLPS., 492 (1): 250-261, DOI: 10.1016/j.gene.2011.10.020.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S., 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods., 28 (10): 2731-2739, DOI: 10.1093/molbev/msr 121.
Tanaka, K. J., Kawamura, H., Matsugu, H., and Nishikata, T., 2000. An ascidian glycine-rich RNA binding protein is not induced by temperature stress but is expressed under a genetic program during embryogenesis., 243 (1-2): 207- 214, DOI: 10.1016/s0378-1119(99)00567-3.
Tate, P. H., and Bird, A. P., 1993. Effects of DNA methylation on DNA-binding proteins and gene expression., 3 (2): 226-231.
Tong, G., Endersfelder, S., Rosenthal, L. M., Wollersheim, S., Sauer, I. M., Buhrer, C., Berger, F., and Schmitt, K. R. L., 2013. Effects of moderate and deep hypothermia on RNA- binding proteins RBM3 and CIRP expressions in murine hippocampal brain slices., 15 (04): 74-84, DOI: 10.1016/j.brainres.2013.01.041.
Uochi, T., and Asashima, M., 1998.(homolog of cold-inducible RNA-binding protein) is expressed tran- siently in developing pronephros and neural tissue., 211 (2): 245-250, DOI: 10.1016/s0378-1119(98)00102-4.
van Venrooy, S., Fichtner, D., Kunz, M., Wedlich, D., and Gradl, D., 2008. Cold-inducible RNA binding protein (CIRP), a novel XTcf-3 specific target gene regulates neural development in., (8): 77, DOI: 10.1186/1471-213x-8-77.
Wellmann, S., Buhrer, C., Moderegger, E., Zelmer, A., Kirschner, R., Koehne, P., Fujita, J., and Seeger, K., 2004. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism., 117 (Pt 9): 1785-1794, DOI: 10.1242/jcs.01026.
Wu, X., Wang, Z., Jiang, J., Gao, J., Wang, J., Zhou, X., and Zhang, Q., 2014. Cloning, expression promoter analysis of vasa gene in Japanese flounder ()..,, (167): 41-50, DOI: 10.1016/j. cbpb.2013.06.004.
Xia, Z., Zheng, X., Zheng, H., Liu, X., Yang, Z., and Wang, X., 2012. Cold-inducible RNA-binding protein (CIRP) regulates target mRNA stabilization in the mouse testis., 586 (19): 3299-3308, DOI: 10.1016/j.febslet.2012.07.004.
Xue, J. H., Nonoguchi, K., Fukumoto, M., Sato, T., Nishiyama, H., Higashitsuji, H., Itoh, K., and Fujita, J., 1999. Effects of ischemia and H2O2 on the cold stress protein CIRP expression in rat neuronal cells., 27 (11-12): 1238-1244, DOI: 10.1016/s0891-5849 (99)00158-6.
Yamamoto, E., 1999. Studies on sex-manipulation and production of cloned populations in hirame,(Temminck et Schlegel)., 173 (1-4): 235-246, DOI: http://dx.doi.org/10.1016/S0044-8486(98) 00448-7.
Yang, C. L., and Carrier, F., 2001. The UV-inducible RNA- binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response., 276 (50): 47277-47284, DOI: 10.1074/jbc.M 105396200.
Zhong, Q., Zhang, Q., Wang, Z., Qi, J., Chen, Y., Li, S., Sun, Y., Li, C., and Lan, X., 2008. Expression profiling and validation of potential reference genes duringembryogenesis.(), 10 (3): 310-318, DOI: 10.1007/s10126-007-9064-7.
Zhou, K. W., Zheng, X. M., Yang, Z. W., Zhang, L., and Chen, H. D., 2009. Overexpression of CIRP may reduce testicular damage induced by cryptorchidism., 32 (2): E103-E111.
(Edited by Ji Dechun)
DOI 10.1007/s11802-015-2622-0
ISSN 1672-5182, 2015 14 (1): 161-170
? Ocean University of China, Science Press and Springer-Verlag Berlin Heidelberg 2015
(March 11, 2014; revised May 19, 2014; accepted June 12, 2014)
* Corresponding author. Tel: 0086-532-82031806 E-mail: qzhang@ouc.edu.cn
 Journal of Ocean University of China2015年1期
Journal of Ocean University of China2015年1期
- Journal of Ocean University of China的其它文章
- A Comparative Study of CART and PTM for Modelling Water Age
- Numerical Study of the Secondary Circulations in Rip Current Systems
- Effect of Anthracene on the Interaction Between Platymonashelgolandica var. tsingtaoensis and Heterosigma akashiwoin Laboratory Cultures
- Application of Numerical Simulation for Analysis of Sinking Characteristics of Purse Seine
- The Structure and Formation Mechanism of a Sea Fog Event over the Yellow Sea
- Effects of the Amplitude and Frequency of Salinity Fluctuation on the Body Composition and Energy Budget of Juvenile Tongue Sole (Cynoglossus semilaevis)
