The Olig family affects central nervous system development and disease
Botao Tan, Jing Yu, Ying Yin, Gongwei Jia, Wei Jiang, Lehua Yu
Department of Rehabilitation Medicine, Second Af fi liated Hospital, Chongqing Medical University, Chongqing, China
The Olig family affects central nervous system development and disease
Botao Tan, Jing Yu, Ying Yin, Gongwei Jia, Wei Jiang, Lehua Yu
Department of Rehabilitation Medicine, Second Af fi liated Hospital, Chongqing Medical University, Chongqing, China
Neural cell differentiation and maturation is a critical step during central nervous system development. The oligodendrocyte transcription family (Olig family) is known to be an important factor in regulating neural cell differentiation. Because of this, the Olig family also affects acute and chronic central nervous system diseases, including brain injury, multiple sclerosis, and even gliomas. Improved understanding about the functions of the Olig family in central nervous system development and disease will greatly aid novel breakthroughs in central nervous system diseases. This review investigates the role of the Olig family in central nervous system development and related diseases.
nerve regeneration; brain injury; spinal cord injury; review; Olig family; oligodendrocytes; astrocytes; central nervous system disease; demyelination; development; differentiation; NSFC grant; neural regeneration
Funding: This study was supported by grants from the National Natural Science Foundation of China, No. 81171859; the Natural Science Foundation of Chongqing, No. cstc2012jjA10058; and the Chongqing Health Bureau Project, No. 2011-2-172.
Tan BT, Yu J,Yin Y, Jia GW, Jiang W, Yu LH. The Olig family affects central nervous system development and disease. Neural Regen Res. 2014;9(3):329-336.
Introduction
The oligodendrocyte transcription family (Olig family) is widely expressed in the central nervous system of various mammals, and plays a critical role in central nervous system development by controlling differentiation and maturation of oligodendrocytes, motor neurons and astrocytes[1-3]. Moreover, accumulating evidence demonstrates Olig family participation in many central nervous system diseases[4-5]. Therefore, based on current literature, we examine the role of the Olig family in central nervous system development and related diseases.
Overview of the Olig famliy
The Olig family consists of three members, Olig1, Olig2 and Olig3, all belonging to the basic helix-loop-helix (bHLH) transcription factor family[6-8]. Olig, Mash, Math and Neurogenin are activating members of the bHLH transcription factor family, and are antagonized by Hes, which acts as an inhibitor within this large family. Activator- and inhibitor-type bHLH genes regulate each other to enable a small number of neural stem cells to differentiate while maintaining others in an undifferentiated state[1]. Olig3 was first discovered in 2000. Olig3 was not initially thought to be expressed in the central nervous system. However, its expression was soon con fi rmed[8]. The function of the Olig family in central nervous system development and disease has gradually been determined (Figure 1).
Both Olig1 and Olig2 map in close proximity on chromosome 16 in rats, the syntenic region to human 21q22. Olig3 maps to chromosome 10 in rats, syntenic to human 6q24. The main function of Olig1 is promotion of oligodendrocyte maturity, and later, myelin formation. Oligodendrocyte precursor cells are generated in Olig1 single mutant mice, but show delayed maturity. Furthermore, during central nervous system development, Olig1 assists Olig2 in formation of the motor neuron progenitor domain (pMN), the area responsible for generation of motor neurons and oligodendrocytes)[9-10]. Olig2 regulates oligodendrocyte and motor neuron differentiation by controlling ventral neuroectodermal precursor cell development. Subsequently, motor neurons and oligodendrocytes are not produced in the spinal cord of Olig2 single mutant mice. In Olig1/2 double-mutant mice, motor neurons are largely absent and oligodendrocyte differentiation abolished[11]. Lineage tracing experiments in Olig1/2 knockout mice, show that pMN progenitors generate V2 interneurons and astrocytes instead[12]. Olig3 is expressed in progenitor cells of class A (dI1–dI3) neurons, and is also important for development of different types of interneurons. Development of class A neurons is impaired in Olig3 mutant mice, with dI1 neurons generated in reduced numbers, and dI2 and dI3 neurons misspeci fi ed, assuming the identity of class B neurons[13-15]. It is now known that Olig family members do not act independently but interact with many other transcription factors, thereby exerting precise control during centralnervous system development. Further evidence indicates the Olig family also participates in central nervous system disease as well.
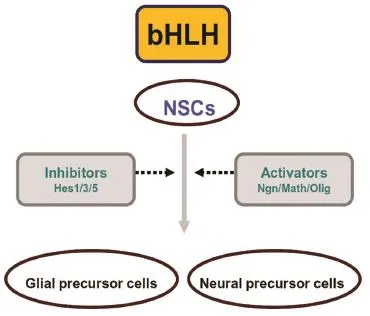
Figure 1 The basic helix-loop-helix (bHLH) family regulates the early differentiation stage of neural stem cells.
The Olig famliy and central nervous system development
The Olig family is present in many vertebrates, including humans, rodents, chickens, and zebra fi sh. Over the years, many studies have been performed to determine the relationship between the Olig family and central nervous system development, using technologies that include gene knock out/in, lineage analysis, and gene transfection.
The Olig family and oligodendrocyte differentiation
Olig1 and Olig2 were fi rst identi fi ed as transcription factors responsible for neural stem cell differentiation into oligodendrocytes, hence the name. Knocking out Olig1 and Olig2 individually or together, impacts upon oligodendrocyte differentiation and maturation, suggesting they functionally overlap in the central nervous system[9-10]. In fact, Olig2 is more likely an early stage factor for oligodendrocyte precursor cells, while Olig1 appears to play a critical role in late stage oligodendrocyte maturation and myelin formation[16]. The nuclear/ cytoplasmic transition of Olig1 and Olig2 is one of the main mechanisms behind their function. Nearly all of Olig1 moves from the nucleus to the cytoplasm within 2 weeks after birth, at which point, oligodendrocyte progenitor cells differentiate into myelin basic protein-positive oligodendrocytes. It has also been shown that Olig1 is located within the nucleus at early stages of remyelinating lesions, and in remyelinating cerebral lesions in postmortem brain tissue from patients with multiple sclerosis[17]. Olig2 remains in the nucleus during oligodendrocyte differentiation and maturation.
The mechanism by which Olig1/2 promotes oligodendrocyte differentiation and maturation remains unclear. However, it is known that they do not work independently, but accompany many other factors, for example, sonic hedgehog (Shh), Sox10, and Nkx2.2[18-20]. Shh is expressed in the embryonic fl oor plate, instructing formation of transcription factor domains along the ventrodorsal axis of the spinal cord[6]. In zebrafish, treatment with Shh inhibitors or Notch blockers, results in reduced or absent Olig1 and Olig2 expression. Although these two factors are necessary during early oligodendrocyte progenitor cell differentiation, levels of both must reduce during the maturation stage[20]. Jagged2, a Notch ligand, is regulated by Shh and its extinction is critical for generation of oligodendrocyte progenitor cells. Overexpression of Olig1 and Olig2 does not rescue loss of Shh signal, suggesting other factors in addition to Olig1 and Olig2, are necessary for oligodendrocyte development[21]. Olig1 physically associates with Sox10, another oligodendrocyte-associated transcription factor, to active myelin basic protein expression in zebra fi sh[19]. Speci fi cally, they work via a conserved (almost homologous with Olig2 in mouse) DNA sequence in the myelin basic protein gene promoter region. The Olig2 and U2 (a conserved Sox10 regulatory region) complex is a direct activator of certain oligodendrocyte-related proteins[22]. Nkx2.2 was discovered as another factor involved in most of the oligodendrocyte differentiation process. Myelin basic protein and proteolipid protein expression (both myelin related proteins) are dramatically decreased in Nkx2.2-null oligodendrocytes[18]. Nkx2.2 acts with Olig2 as a co-activator of myelin genes during oligodendrocyte development. Before the end of myelination, Wnt/beta-catenin signaling and bone morphogenetic protein show a trend towards down-regulation, as both are blocked by Smad7, a myelination positive protein[23-24]. Thus, bone morphogenetic protein plays a negative role during oligodendrocyte differentiation and remyelination. Overexpression of both Olig1 and Olig2, causes differentiation of oligodendrocyte progenitor cells to oligodendrocytes by inhibiting bone morphogenetic proteins 2 and 4, suggesting Olig1 and Olig2 can be used to treat demyelinating diseases of the central nervous system[24]. The ATP-dependent chromatin-remodeling enzyme, Smarca4/Brg1, is necessary and suf fi cient to initiate and promote oligodendrocyte lineage development. Functional analyses of Smarca4/Brg1 and Olig2 co-occupied chromatin epigenetic marking, uncovered stage-speci fi c elements that predict sets of transcriptional regulators controlling oligodendrocyte differentiation. Regulation of the Smarca4/Brg1-dependent chromatin-remodeling complex by Olig2, coupled with other transcription factor linked chromatin modi fi cations, is critical for precise initiation and establishment of oligodendrocyte differentiation and maturation[25](Figure 2).
The Olig family and neuronal differentiation
While Olig2 regulates development of ventral spinal motor neurons, Olig3 ensures orderly dorsal interneuron differentiation[4,13].
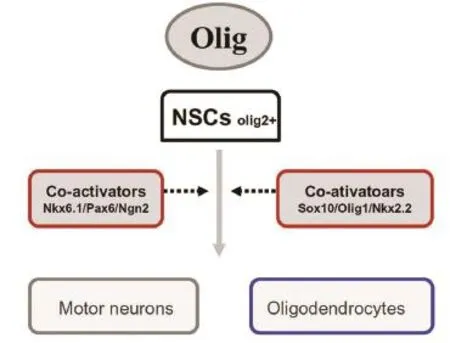
Figure 2 The Olig family regulates motor neuron and oligodendrocyte differentiation.
Under the control of many growth genes during early central nervous system development, the ventral neuroepithelium is divided into five distinct domains (p0-p3 and pMN). Each domain generates a corresponding class of neurons, containing V0-V3 interneurons and motor neurons[26]. Olig2 is uniquely expressed in pMN cells, enabling them to differentiate into motor neurons through cooperation with other factors[27-28]. Both oligodendrocytes and motor neurons are derived from the pMN domain. Olig1/2 double-mutant mice lack spinal motor neurons and oligodendrocytes, due to transformation of pMN cells into progenitor V2 interneurons and astrocytes[9]. Spinal cord pMN cells give rise to motor neurons in the first wave of differentiation, and then to oligodendrocytes. Subsequently, Olig2 and its co-working transcription factors gradually diminish in motor neuron progenitors, accompanied by the appearance of several speci fi c motor markers, such as Hb9 and islet1/2[21,29].
Olig2 is crucial for both oligodendrocyte and motor neuron differentiation in many loss and gain of function studies. However, the exact conditions facilitating motor neuron development are not known. Throughout the process, Olig2 must interact with other transcription factors to fulfill its role. Except for certain common factors with oligodendrocyte maturation, motor neuron differentiation requires timely expression of specific genes, for example, Nkx6.1 and Ngn2. Early studies found Nkx6.1 expressed in the ventral neural tube, and its activity necessary for motor neuron generation. In addition, Nkx6.1 controls oligodendrocyte development in the spinal cord, and plays an instructive role in controlling both motor neuron and oligodendrocyte speci fi cation through regulation of Olig2 expression at different stages[30]. Olig2 and Ngn2 are co-expressed in specific regions of the central nervous system that generate motor neurons. Moreover, aberrant expression of Olig2 and Ngn2 induces many neural stem cells to express motor neuron markers[27-28,31], indicating the complex regulates Olig2 and Ngn2 interactions during motor neuron differentiation. It was further shown that when Olig2 primes pMN cells, generating motor neurons, it must trigger Ngn2 and Lhx3 expression. Olig2 may serve the function of preventing premature motor neuron development, and Ngn2 a counter role, by releasing a subset of differentiating Olig2+progenitors and activating post-mitotic motor neuron makers[31-32]. The Olig2 phosphorylation state may be another critical step in promotion of oligodendrocyte or motor neuron differentiation. It was recently determined that Olig2 phosphorylation at serine 147, increases efficiency of Olig2-Ngn2 heterodimer formation by reducing the amount of Ngn2 available for activating motor neuron-speci fi c genes, thereby contributing to neuron-oligodendrocyte fate switch[32].
In 2001, using whole-mount in situ hybridization, Japanese researchers found Olig3 expressed in different progenitor types of the embryonic central nervous system[8,33]. Olig3 is detected in the dorsal neural tube from the midbrain/hindbrain boundary to the spinal cord, indicating that Olig3 may function in central nervous system development.
Olig3 was thought to play an important role in fate specification and differentiation of various neurons, especially dorsal spinal cord neurons[13-15,34-35]. In zebra fi sh, early loss of function studies showed that Olig3 affects establishment of the neural crest-lateral neural plate boundary that is necessary for spinal cord development. Short-term lineage analysis of dorsally derived Olig3+cells in the developing spinal cord, found they contributed to dorsal midline neurons and commissural interneurons at intermediate and ventral levels. Some cells expressed Islet1/2, Math1, and Brn3a, markers for dorsal interneurons or sensory neurons at the alar-plate[14]. Further studies con fi rmed requirement of Olig3 for correct development of class A neuronal subtypes in the dorsal spinal cord and hindbrain[15,35]. In Olig3 mutant mice, class A neuronal development is impaired[13]. Consequently, the brainstem solitary nucleus tract does not form, and animals show absent or smaller inferior olivary nuclei[15]. During embryonic stages, Olig3 is co-expressed with other makers (including Islet1/2, Ngn1, Mash1, Math1, and Brn3a), and regulated by the Notch and Wnt signaling pathways[34-35]. Therefore, just as with Olig2 promotion of motor neuron and oligodendrocyte development, Olig3 never acts alone. The mechanism by which Olig3 regulates differentiation of various central nervous system cells also requires further investigation.
The Olig family and astrocyte differentiation
The Olig family is not only essential for oligodendrocyte and motor neuron development, but also astrocyte development[2]. Use of a long-term tracking system confirmed that Olig2+precursor cells in pMN areas differentiate into motor neurons and oligodendrocytes, and that a portion of astrocytes distributed in the ventral spinal cord or preven-tricular ependymal cells, are also derived from these pMN area cells[3]. It was observed that most telencephalic Olig2+cells differentiated into glial cells (including astrocytes and oligodendrocytes) in late embryonic stages[36]. In Olig2 gene knockout mice, white matter astrocytes show maturation dif fi culties. Therefore, Olig2 may in fl uence proper astrocyte development as well. However, in vitro studies found Olig 2 mutant neural progenitor cells show a trend towards astrocyte differentiation. During astrocyte differentiation in neonatal mice, Olig2 is transiently expressed in immature astrocytes, gradually diminishing with later astrocyte maturation. Other research shows Olig2 in the nucleus of neural stem cells, blocks ciliary neurotrophic factor-mediated astrocyte differentiation by inhibiting both STAT3 and p300 expression[38]. When cultured, neural stem cells differentiate into glial fi brillary acidic protein (GFAP)-positive astrocytes, and nuclear Olig2 disappears and is instead detected in the cytoplasm. Cytoplasmic Olig2+cells are also GFAP (an astrocyte-specific marker)-positive, whereas nuclear Olig2+cells mainly differentiate into oligodendrocytes[2]. Rapid removal of Olig2 from the cell nucleus may be a critical step for astrocyte differentiation.
The Olig famliy in central nervous disease
As the Olig family is known to be important for varied cell types, especially oligodendrocytes and motor neurons, it is easy to speculate that the Olig family may participate in certain restorative processes. Many studies have shown the Olig family (particularly Olig1 and Olig2 members), promotes restoration of the demyelinated central nervous system and may contribute to neural regeneration[4-5,39]. Additionally, many studies have closely examined the relationship between Olig2 alone, and central nervous system gliomas[40-42].
The Olig family and demyelination
Multiple sclerosis is the most commonly studied demyelination disease, and always causes neurological disability in humans. Relapsing demyelination (oligodendrocyte loss) and ensuing axonal deterioration are the primary pathological mechanisms of multiple sclerosis[43]. Some oligodendrocyte progenitor cells have the ability to proliferate, migrate, and differentiate into myelin-forming oligodendrocytes, but they fail to lead remyelination because of insufficient number and efficiency of the oligodendrocyte progenitor cells[44-45]. Therefore, understanding the mechanisms underlying remyelination is a valuable potential therapeutic target for central nervous system diseases, such as multiple sclerosis. Progenitor cells preferentially expressing Olig2, differentiate into oligodendrocytes and are observed in cuprizone-induced demyelinated lesions, indicating Olig1 and Olig2 are needed not only in oligodendrocyte differentiation, but also remyelination[18].
Expression of the Olig family (mainly Olig1 and Olig2) and other myelin-related factors (e.g., Nkx2.2) occurs at higher levels in the ethidium bromide induced demyelination model, compared with the normal condition. This has been confirmed by Talbott et al.[46], in a spinal cord demyelination model. Given the decisive role of the Olig gene in central nervous system development, the expression change in oligodendrocyte progenitor cells may aid differentiation into remyelinated oligodendrocytes[47]. The fate of Olig2+progenitors in demyelinated adult mice has been investigated using long-term tracking technology, and shows that most Olig2+progenitors proliferate and differentiate into oligodendrocytes more ef fi ciently compared with normal mice[48]. Many studies have con fi rmed that Olig1 and/ or Olig2 up-regulation in stem or progenitor cells leads to obvious myelination[39,49-51]. Bioluminescence imaging was used as a noninvasive means to measure the fate of neural stem cells isolated from luciferase-green fluorescent protein (GFP)-actin transgenic mice, that underwent Olig2 or GFP (as control) gene interference after implantation in the corpus callosum of cuprizone-induced demyelinated mice. It was shown that the majority of surviving Olig2 overexpressing neural stem cells differentiated into oligodendrocytes, and finally became remyelinated axons[49]. Neural stem cell survival rates were signi fi cantly higher in the Olig2-neural stem cell group than in the GFP-neural stem cell group, indicating that if implanted neural stem cells did not undergo differentiation, they rapidly died. Electron microscopy has shown Olig2-neural stem cells exhibit different phases of myelin formation. Similar research obtained the same outcomes. Transplantation of human neural stem cells transfected with the Olig2 gene, enhanced white matter myelination in a rat spinal cord injury model, and ultimately improved hindlimb locomotor function[50]. Injection of Olig1 and Olig2 recombinant retroviruses into injured rat spinal cord had strikingly similar results[51]. Doxycycline-inducible Olig2 expression in nestin-expressing progenitor cells in the mouse central nervous system, leads to increased and precocious myelination[39].
Analysis of oligodendrocyte maturation in Olig1 knockout mice after demyelination demonstrates these progenitors cannot complete differentiation[47]. Activation of the Olig gene in demyelination is not accompanied with increased Shh levels, the important trigger for the Olig gene during development, suggesting the Olig family does not perform precisely the same role in repair processes and oligodendrocyte development[18]. Similar research has shown that besides Olig1 and Olig2, other factors (including NG2, Nkx2.2, Sox10, and astrocytes) are necessary for the remyelination process. Moreover, according to Arnett’s research, Olig1 subcellular localization (either inside or outside the nucleus) is essential for oligodendrocyte maturation[17]. There are many factors involved in remyelination, yet the Olig gene takes a central position. Problems include: 1) what is the optimal strategy for Olig gene overexpression? 2) what is the optimal time point to start gene interference, and as it is not feasible to maintain sustained high Olig gene expression, when must re-expression be stopped? and 3) how can the safety of transferring Olig gene therapies to patients be ensured? In retrovirus injection models, Olig2 is involved in tumor transformation. If these issues can be solved, it may be pos-sible to predict and prevent some demyelinating diseases by regulating Olig gene expression.
The Olig family and central nervous system injury
A combination of factors is responsible for the lack of neural regeneration and minimal functional recovery generally observed after central nervous system injury, with the most striking cases being spinal cord injury and cerebral infarction[52]. Endogenous neural stem cells in the central nervous system are activated to generate more daughter cells in response to injury, but barely cause recovery, suggesting that it is not enough to recruit and activate these stem cells[5].
Many studies have shown that most activated endogenous neural stem cells cannot complete differentiation, and even die, dependent on the transplantation strategies[53-55]. The majority of surviving stem cells give rise to astrocytes, and thereby finally block restoration by glial scar formation[56]. By providing neurotrophic support and modulating the infl ammatory response, the functional bene fi ts of transplanting stem cells without neuron or glial cell generation has been reported[57]. The original aim for activating stem cells is to produce neurons and other cells, a small number of which are generated[58]. Therefore, it is attractive to consider ways to facilitate stem cell differentiation into neurons and oligodendrocytes. Stimulation with neurotrophic factors (e.g., epidermal or fibroblast growth factors), ensures stem cells have a greater chance of differentiating into neurons[59-60]. Endogenous neural stem cell differentiation depends on expression of development-related factors. The Olig family has been shown to regulate different types of neurons in the developing central nervous system, but its exact role in central nervous system injury has not been clearly elucidated. Many transcription factors (including the Olig family) show altered expression patterns after central nervous system injury and other acute/chronic diseases[60-61]. Olig1 and Olig3 expression increases strongly with the BrdU state, while Olig2 is mainly expressed in neuronal precursors. It appears that the Olig family, and most importantly Olig2, is broadly engaged in neural repair process after central nervous system injury[53,62]. Intracellular Olig2 localization is associated with the differentiation direction of NG2+progenitors after acute brain injury.
We have not yet considered how the mammalian spinal cord regenerates motor neurons after injury[63]. In adult zebrafish, the situation appears different[64]. It has been demonstrated that proliferating Olig2+ependymal radial glial progenitor cells switch from a gliogenic phenotype to functional motor neurons. This renewal process is impaired by blocking Shh signaling. It is apparent that Olig2 up-regulation is beneficial for neural regeneration in zebrafish. Moreover, a large number of studies on the Olig2 transgene in spinal cord injury models, reach the same outcome of improved locomotor recovery in rodents, although its mechanisms may be associated with oligodendrocyte regeneration[51,65]. Buffo et al.[62]found that continuously high Olig2 expression represses neurogenesis in response to brain injury, as antagonizing Olig2 function results in a signi fi cant number of infected cells generating immature neurons, that are not observed after injection with a control virus. Therefore, down-regulating Olig2 at the appropriate time may be crucial to differentiation of neuronal lineage cells, similar to the developing spinal cord. This may explain the lack of definite motor neuron regeneration in studies examining up-regulated Olig2 expression[66]. Further studies are needed to focus on appropriate temporal expression patterns of the Olig family, and other related transcription factors, after central nervous system injury for optimal functional neural regeneration.
The Olig family and central nervous system tumors
Gliomas are classified as astrocytomas, oligodendrogliomas, or mixed oligoastrocytomas, based solely on their histological resemblance to astrocytes or oligodendrocytes. Most of these glial derived brain tumors cause severe neurological dysfunction, and even death. Knowledge of glia tumor etiology, nosogenesis, and early diagnosis, is bene fi cial for treatment.
Gliomas are thought to arise from progenitor/stem cells[67]. Considering the effects of the Olig family in normal central nervous system development, we speculate that the Olig gene may be expressed in certain gliomas. Previous studies have demonstrated high levels of Olig1 and Olig2 mRNA in oligodendrogliomas[40,68]. However, at the protein level, the Olig family is not restricted to oligodendrogliomas, and is also expressed in most diffuse gliomas, including astrocytomas, oligodendrogliomas, and oligo-astrocytomas[41,68-69]. Expression patterns of the Olig family in different gliomas is unpredictable. In some cases, Olig gene expression in astrocytomas even matched levels in oligodendroglial tumors. Otero and colleagues tested 90 non-oligodendroglial pediatric gliomas, and found Olig2 expression varied from very low levels in ependymomas to high levels in pilocytic astrocytomas and diffuse-type astrocytic tumors[70]. Thus, the Olig family is not appropriate for sole diagnosis of oligodendrogliomas. Nevertheless, compared with neurocytomas and glioblastomas, the Olig gene is largely expressed in gliomas, indicating it may participate in glia tumorigenesis mechanisms[71-72]. Olig2 is detected in a subpopulation of type B and type C (mainly type C) cells in the adult rodent central nervous system[73-74]. These Olig2+type C cells undergo proliferation and glioma-like growth after exposure to epidermal or platelet-derived growth factors[75].
Both epidermal and platelet-derived growth factors are known glioma-related mitogens[76]. Glial progenitor cells in the adult human brain show potential as the origin for human gliomas. Quantitative analysis of these mitotic cells tends to implicate Olig2 in glioma histogenesis and biology[67]. Accordingly, it is clear that Olig2 is likely important for glioma oncogenesis.
Experiments have shown that cultured glioma formation from neurospheres, lose their tumorigenic capacity after Olig2 null, but not Olig1[42,77]. In human glioblastoma samples, most CD133+or Ki67+cells express Olig2, indicating Olig2 may regulate proliferation of tumorigenic neuro-spheres. Infection of Olig1/2 knockout EGFRvIII (tumorigenic) neurospheres with Olig2 retrovirus, completely restores cells to the morphology and proliferation of wild-type EGFRvIII neurospheres. Similarly, Olig2 is essential for cell proliferation and tumorigenic potential of platelet-derived growth factor-induced oligodendrogliomas. RNAi of Olig2 resulted in oligodendrogliomas with decreased spreading ability in a platelet-derived growth factor-B derived human cell line[72]. Using mass spectroscopy, phosphorylation state-speci fi c antibodies, and multiple gain and loss studies, American researchers have found that phosphorylation of a conserved triple serine motif of Olig2 serves the role of its promitotic functions in human glioma neurosphere cultures and in a murine model of primary glioma[77-78]. It was further discovered that p53 acetylation is suppressed by Olig2 in neural progenitors and malignant gliomas. Posttranslational modification (phosphorylation and acetylation) of p53 transactivates downstream factors, leading to either growth arrest or cell death of target tumor cells. Thus, the reason why high-grade gliomas, which have an intact p53 gene, are insensitive to radiation and genotoxic drugs becomes clearer.
The molecular mechanisms of the Olig family in controlling central nervous system development and neoplastic disease, highlights therapeutic opportunities for related cancers. The oppositional relationship of phosphorylated Olig2 with the p53 signal, suggests that small molecule inhibitors of the Olig2 phosphorylation state protein kinase may have practical applications in glioma therapy. How to make these inhibitors work and not affect Olig2 expression in non-glioma regions simultaneously, will be a key and dif fi cult point in future therapy strategies.
Conclusion
From our above discussion, we are better acquainted with the Olig family. They all act as critical factors in selecting neural cell subtypes. Olig1 affects oligodendrocyte differentiation and maturation, Olig3 takes part in formation of interneurons in the dorsal spinal cord and hindbrain, and Olig2 plays an important role in regulating the appearance and development of both motor neurons and oligodendrocytes.
Consequently, central nervous system functions of the Olig family members, imply an extensive relationship between the Olig family and central nervous system disorders. To date, multiple sclerosis, central nervous system injury, and various gliomas, all exhibit strong correlation with the Olig family. Although the Olig family never acts independently and a number of other factors show equal status in controlling correct neuronal cell differentiation, altering expression levels of Olig family members, often brings therapeutic effects in the above mentioned central nervous system diseases. Olig1/2 overexpression promotes oligodendrocyte regeneration and further remyelination, in lesions of drug-induced demyelination animal models. Neural precursor cells are able to differentiate into motor neurons in vitro following transfection of Olig2 overexpression vectors. Olig2 maintains high expression levels in certain gliomas, for example glioblastomas, and blocks the p53 gene, which fi nally leads to drug resistance as well as tumor progression. All these fi ndings give further hope for dealing with intractable neural regeneration problems and two other neurological puzzles.
In summary, the Olig family and its regulatory pathways need to be further examined in future research. This will provide novel avenues in fi nding new treatment strategies for multiple sclerosis and other central nervous system diseases.
Author contributions:Yu LH was responsible for study design and implementation. Tan BT prepared the manuscript. Yu J and Yin Y assisted in data collection and analysis. Jia GW and Jiang W assisted in manuscript preparation and revision. All authors approved the final version of the manuscript.
Con fl icts of interest:None declared.
[1] Kageyama R, Ohtsuka T, Hatakeyama J, et al. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343-348.
[2] Zhao JW, Raha-Chowdhury R, Fawcett JW, et al. Astrocytes and oligodendrocytes can be generated from NG2+progenitors after acute brain injury: intracellular loalization of oligodendrocyte transcription factor 2 is associated with their fate choice. Eur J Neurosci. 2009;29:1853-1869.
[3] Masahira N, Takebayashi H, Ono K, et al. Olig2-positive progenitors in the embryonic spinal cord give rise not only to motoneurons and oligodendrocytes, but also to a subset of astrocytes and ependymal cells. Dev Biol. 2006;293:358-369.
[4] Ligon KL, Fancy SP, Franklin RJ, et al. Olig gene function in CNS development and disease. Glia. 2006;54:1-10.
[5] Barreiro-Iglesias A. Targeting ependymal stem cells in vivo as a non-invasive therapy for spinal cord injury. Dis Model Mech. 2010;3:667-668.
[6] Lu QR, Yuk D, Alberta JA, et al. Sonic hedge-hog--regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317-329.
[7] Zhou Q, Wang S, Anderson DJ. Identi fi cation of a novel family of oligodendrocyte lineage-speci fi c basic he-lix-loop-helix transcription factors. Neuron. 2000;25:331-343.
[8] Takebayashi H, Yoshida S, Sugimori M, et al. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identi fication of a new member, Olig3. Mech Dev. 2000;99:143-148.
[9] Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype speci fi cation. Cell. 2002;109:61-73.
[10] Lu QR, Sun T, Zhu Z, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75-86.
[11] Takebayashi H, Nabeshima Y, Yoshida S, et al. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157-1163.
[12] Sun T, Echelard Y, Lu R, et al. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr Biol. 2001;11:1413-1420.
[13] Muller T, Anlag K, Wildner H, et al. The bHLH factor Olig3 coordinates the specification of dorsal neurons in the spinal cord. Genes Dev. 2005;19:733-743.
[14] Ding L, Takebayashi H, Watanabe K, et al. Short-term lineage analysis of dorsally derived Olig3 cells in the developing spinal cord. Dev Dyn. 2005;234:622-632.
[15] Storm R, Cholewa-Waclaw J, Reuter K, et al. The bHLH transcription factor Olig3 marks the dorsal neuroepithelium of the hindbrain and is essential for the development of brainstem nuclei. Development. 2009;136:295-305.
[16] Othman A, Frim DM, Polak P, et al. Olig1 is expressed in human oligodendrocytes during maturation and regeneration. Glia. 2011;59:914-926.
[17] Arnett HA, Fancy SP, Alberta JA, et al. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111-2115.
[18] Fancy SP, Zhao C, Franklin RJ. Increased expression of Nkx2.2 and Olig2 identi fi es reactive oligodendrocyte progenitor cells responding to demyelination in the adult CNS. Mol Cell Neurosci. 2004;27:247-254.
[19] Li H, Lu Y, Smith HK, et al. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375-14382.
[20] Schebesta M, Serluca FC. olig1 Expression identifies developing oligodendrocytes in zebrafish and requires hedgehog and notch signaling. Dev Dyn. 2009;238:887-898.
[21] Rabadan MA, Cayuso J, Le Dreau G, et al. Jagged2 controls the generation of motor neuron and oligodendrocyte progenitors in the ventral spinal cord. Cell Death Differ. 2012;19:209-219.
[22] Kuspert M, Hammer A, Bosl MR, et al. Olig2 regulates Sox10 expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. Nucleic Acids Res. 2011;39:1280-1293.
[23] Weng Q, Chen Y, Wang H, et al. Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron. 2012;73:713-728.
[24] Cheng X, Wang Y, He Q, et al. Bone morphogenetic protein signaling and olig1/2 interact to regulate the differentiation and maturation of adult oligodendrocyte precursor cells. Stem Cells. 2007;25:3204-3214.
[25] Yu Y, Chen Y, Kim B, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248-261.
[26] Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell speci fi cation. Nature. 2010;468:214-222.
[27] Mizuguchi R, Sugimori M, Takebayashi H, et al. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757-771.
[28] Lee SK, Lee B, Ruiz EC, et al. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282-294.
[29] Thaler JP, Lee SK, Jurata LW, et al. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237-249.
[30] Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773-789.
[31] Lee SK, Jurata LW, Funahashi J, et al. Analysis of embryonic motoneuron gene regulation: derepression of general activators function in concert with enhancer factors. Development. 2004;131:3295-3306.
[32] Li H, de Faria JP, Andrew P, et al. Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron. 2011;69:918-929.
[33] Takebayashi H, Ohtsuki T, Uchida T, et al. Non-overlapping expression of Olig3 and Olig2 in the embryonic neural tube. Mech Dev. 2002;113:169-174.
[34] Liu Z, Li H, Hu X, et al. Control of precerebellar neuron development by Olig3 bHLH transcription factor. J Neurosci. 2008; 28:10124-10133.
[35] Zechner D, Muller T, Wende H, et al. Bmp and Wnt/beta-catenin signals control expression of the transcription factor Olig3 and the speci fi cation of spinal cord neurons. Dev Biol. 2007;303:181-190.
[36] Ono K, Takebayashi H, Ikeda K, et al. Regional- and temporal-dependent changes in the differentiation of Olig2 progenitors in the forebrain, and the impact on astrocyte development in the dorsal pallium. Dev Biol. 2008;320:456-468.
[37] Cai J, Chen Y, Cai WH, et al. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134:1887-1899.
[38] Setoguchi T, Kondo T. Nuclear export of OLIG2 in neural stem cells is essential for ciliary neurotrophic factor-induced astrocyte differentiation. J Cell Biol. 2004;166:963-968.
[39] Maire CL, Wegener A, Kerninon C, et al. Gain-of-function of Olig transcription factors enhances oligodendrogenesis and myelination. Stem Cells. 2010;28:1611-1622.
[40] Lu QR, Park JK, Noll E, et al. Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc Natl Acad Sci U S A. 2001;98:10851-10856.
[41] Ohnishi A, Sawa H, Tsuda M, et al. Expression of the oligodendroglial lineage-associated markers Olig1 and Olig2 in different types of human gliomas. J Neuropathol Exp Neurol. 2003;62:1052-1059.
[42] Ligon KL, Huillard E, Mehta S, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503-517.
[43] Compston A, Coles A. Multiple sclerosis. Lancet. 2002;359:1221-1231.
[44] Edgar JM, McLaughlin M, Yool D, et al. Oligodendroglial modulation of fast axonal transport in a mouse model of hereditary spastic paraplegia. J Cell Biol. 2004;166:121-131.
[45] Watanabe M, Hadzic T, Nishiyama A. Transient upregulation of Nkx2.2 expression in oligodendrocyte lineage cells during remyelination. Glia. 2004;46:311-322.
[46] Talbott JF, Loy DN, Liu Y, et al. Endogenous Nkx2.2+/Olig2+ oligodendrocyte precursor cells fail to remyelinate the demyelinated adult rat spinal cord in the absence of astrocytes. Exp Neurol. 2005;192:11-24.
[47] Xin M, Yue T, Ma Z, et al. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354-1365.
[48] Islam MS, Tatsumi K, Okuda H, et al. Olig2-expressing progenitor cells preferentially differentiate into oligodendrocytes in cuprizone-induced demyelinated lesions. Neurochem Int. 2009;54:192-198.
[49] Sher F, van Dam G, Boddeke E, et al. Bioluminescence imaging of Olig2-neural stem cells reveals improved engraftment in a demyelination mouse model. Stem Cells. 2009;27:1582-1591.
[50] Hwang DH, Kim BG, Kim EJ, et al. Transplantation of human neural stem cells transduced with Olig2 transcription factor improves locomotor recovery and enhances myelination in the white matter of rat spinal cord following contusive injury. BMC Neurosci. 2009;10:117.
[51] Kim HM, Hwang DH, Choi JY, et al. Differential and cooperative actions of Olig1 and Olig2 transcription factors on immature proliferating cells after contusive spinal cord injury. Glia. 2011;59:1094-1106.
[52] Barnabe-Heider F, Frisen J. Stem cells for spinal cord repair. Cell Stem Cell. 2008;3:16-24.
[53] Meletis K, Barnabe-Heider F, Carlen M, et al. Spinal cord injury reveals multilineage differentiation of ependymal cells. PLoS Biol. 2008;6:e182.
[54] Horky LL, Galimi F, Gage FH, et al. Fate of endogenous stem/ progenitor cells following spinal cord injury. J Comp Neurol. 2006;498:525-538.
[55]Watt FM, Driskell RR. The therapeutic potential of stem cells. Philos Trans R Soc Lond B Biol Sci. 2010;365:155-163.
[56] Busch SA, Silver J. The role of extracellular matrix in CNS regeneration. Curr Opin Neurobiol. 2007;17:120-127.
[57] Parr AM, Tator CH, Keating A. Bone marrow-derived mesenchymal stromal cells for the repair of central nervous system injury. Bone Marrow Transplant. 2007;40:609-619.
[58] Ronaghi M, Erceg S, Moreno-Manzano V, et al. Challenges of stem cell therapy for spinal cord injury: human embryonic stem cells, endogenous neural stem cells, or induced pluripotent stem cells? Stem Cells. 2010;28:93-99.
[59] Xu Y, Kitada M, Yamaguchi M, et al. Increase in bFGF-responsive neural progenitor population following contusion injury of the adult rodent spinal cord. Neurosci Lett. 2006;397:174-179.
[60] Ohori Y, Yamamoto S, Nagao M, et al. Growth factor treatment and genetic manipulation stimulate neurogenesis and oligodendrogenesis by endogenous neural progenitors in the injured adult spinal cord. J Neurosci. 2006;26:11948-11960.
[61] Tonchev AB, Yamashima T, Sawamoto K, et al. Transcription factor protein expression patterns by neural or neuronal progenitor cells of adult monkey subventricular zone. Neuroscience. 2006;139:1355-1367.
[62] Buffo A, Vosko MR, Erturk D, et al. Expression pattern of the transcription factor Olig2 in response to brain injuries: implications for neuronal repair. Proc Natl Acad Sci U S A. 2005;102:18183-18188.
[63] Reimer MM, Kuscha V, Wyatt C, et al. Sonic hedgehog is a polarized signal for motor neuron regeneration in adult zebrafish. J Neurosci. 2009;29:15073-15082.
[64] Reimer MM, Sorensen I, Kuscha V, et al. Motor neuron regeneration in adult zebra fi sh. J Neurosci. 2008;28:8510-8516.
[65] Hu JG, Shen L, Wang R, et al. Effects of olig2-overexpressing neural stem cells and myelin basic protein-activated T cells on recovery from spinal cord injury. Neurotherapeutics. 2012;9:422-445.
[66] Hack MA, Saghatelyan A, de Chevigny A, et al. Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat Neurosci. 2005;8:865-872.
[67] Rhee W, Ray S, Yokoo H, et al. Quantitative analysis of mitotic Olig2 cells in adult human brain and gliomas: implications for glioma histogenesis and biology. Glia. 2009;57:510-523.
[68] Ligon KL, Alberta JA, Kho AT, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63:499-509.
[69] Azzarelli B, Miravalle L, Vidal R. Immunolocalization of the oligodendrocyte transcription factor 1 (Olig1) in brain tumors. J Neuropathol Exp Neurol. 2004;63:170-179.
[70] Otero JJ, Rowitch D, Vandenberg S. OLIG2 is differentially expressed in pediatric astrocytic and in ependymal neoplasms. J Neurooncol. 2011;104:423-438.
[71] Okada M, Yano H, Hirose Y, et al. Olig2 is useful in the differential diagnosis of oligodendrogliomas and extraventricular neurocytomas. Brain Tumor Pathol. 2011;28:157-161.
[72] Appolloni I, Calzolari F, Barilari M, et al. Antagonistic modulation of gliomagenesis by Pax6 and Olig2 in PDGF-induced oligodendroglioma. Int J Cancer. 2012;131:E1078-1087.
[73] Hack MA, Sugimori M, Lundberg C, et al. Regionalization and fate speci fi cation in neurospheres: the role of Olig2 and Pax6. Mol Cell Neurosci 2004;25:664-678.
[74] Menn B, Garcia-Verdugo JM, Yaschine C, et al. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907-7918.
[75] Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, et al. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187-199.
[76] Calzolari F, Appolloni I, Tutucci E, et al. Tumor progression and oncogene addiction in a PDGF-B-induced model of gliomagenesis. Neoplasia. 2008;10:1373-1382, following 1382.
[77] Mehta S, Huillard E, Kesari S, et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011;19:359-371.
[78] Sun Y, Meijer DH, Alberta JA, et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011;69:906-917.
Copyedited by James R, Haase R, Liu CY, Li X, Li CH, Song LP, Zhao M
10.4103/1673-5374.128232
Lehua Yu, M.D., Ph.D., Department of Rehabilitation Medicine, Second Affiliated Hospital, Chongqing Medical University, Chongqing 400010, China, yulehuadoc@ aliyun.com.
http://www.nrronline.org/
Accepted: 2013-11-15
 中國(guó)神經(jīng)再生研究(英文版)2014年3期
中國(guó)神經(jīng)再生研究(英文版)2014年3期
- 中國(guó)神經(jīng)再生研究(英文版)的其它文章
- Short-term environmental enrichment exposure induces proliferation and maturation of doublecortin-positive cells in the prefrontal cortex
- Special function of nestin+neurons in the medial septum-diagonal band of Broca in adult rats
- Normalization of ventral tegmental area structure following acupuncture in a rat model of heroin relapse
- Acupuncture at Waiguan (SJ5) and sham points in fl uences activation of functional brain areas of ischemic stroke patients: a functional magnetic resonance imaging study
- Can amino-functionalized carbon nanotubes carry functional nerve growth factor?
- Quantitative volumetric analysis of the optic radiation in the normal human brain using diffusion tensor magnetic resonance imaging-based tractography
